Autophagy, immunity, and microbial adaptations
- PMID: 19527881
- PMCID: PMC2720763
- DOI: 10.1016/j.chom.2009.05.016
Autophagy, immunity, and microbial adaptations
Abstract
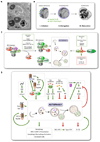
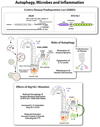
Autophagy adjusts cellular biomass and function in response to diverse stimuli, including infection. Autophagy plays specific roles in shaping immune system development, fueling host innate and adaptive immune responses, and directly controlling intracellular microbes as a cell-autonomous innate defense. As an evolutionary counterpoint, intracellular pathogens have evolved to block autophagic microbicidal defense and subvert host autophagic responses for their survival or growth. The ability of eukaryotic pathogens to deploy their own autophagic machinery may also contribute to microbial pathogenesis. Thus, a complex interplay between autophagy and microbial adaptations against autophagy governs the net outcome of host-microbe encounters.
Figures

References
-
- Alvarez VE, Kosec G, Sant’Anna C, Turk V, Cazzulo JJ, Turk B. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J. Biol. Chem. 2008;283:3454–3464. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

