Differential Spo0A-mediated effects on transcription and replication of the related Bacillus subtilis phages Nf and phi29 explain their different behaviours in vivo
- PMID: 19528067
- PMCID: PMC2731898
- DOI: 10.1093/nar/gkp504
Differential Spo0A-mediated effects on transcription and replication of the related Bacillus subtilis phages Nf and phi29 explain their different behaviours in vivo
Abstract
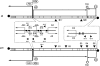
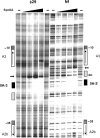
Members of groups 1 (e.g. 29) and 2 (e.g. Nf) of the 29 family of phages infect the spore forming bacterium Bacillus subtilis. Although classified as lytic phages, the lytic cycle of 29 can be suppressed and its genome can become entrapped into the B. subtilis spore. This constitutes an alternative infection strategy that depends on the presence of binding sites for the host-encoded protein Spo0A in the 29 genome. Binding of Spo0A to these sites represses 29 transcription and prevents initiation of DNA replication. Although the Nf genome can also become trapped into B. subtilis spores, in vivo studies showed that its lytic cycle is less susceptible to spo0A-mediated suppression than that of 29. Here we have analysed the molecular mechanism underlying this difference showing that Spo0A differently affects transcription and replication initiation of the genomes of these phages. Thus, whereas Spo0A represses all three main early promoters of 29, it only represses one out of the three equivalent early promoters of Nf. In addition, contrary to 29, Spo0A does not prevent the in vitro initiation of Nf DNA replication. Altogether, the differences in Spo0A-mediated regulation of transcription and replication between 29 and Nf explain their different behaviours in vivo.
Figures






Similar articles
-
Different responses to Spo0A-mediated suppression of the related Bacillus subtilis phages Nf and phi29.Environ Microbiol. 2009 May;11(5):1137-49. doi: 10.1111/j.1462-2920.2008.01845.x. Epub 2009 Jan 15. Environ Microbiol. 2009. PMID: 19207565
-
Spo0A, the key transcriptional regulator for entrance into sporulation, is an inhibitor of DNA replication.EMBO J. 2006 Aug 23;25(16):3890-9. doi: 10.1038/sj.emboj.7601266. Epub 2006 Aug 3. EMBO J. 2006. PMID: 16888621 Free PMC article.
-
Molecular basis for the exploitation of spore formation as survival mechanism by virulent phage phi29.EMBO J. 2005 Oct 19;24(20):3647-57. doi: 10.1038/sj.emboj.7600826. Epub 2005 Sep 29. EMBO J. 2005. PMID: 16193065 Free PMC article.
-
DNA bending and looping in the transcriptional control of bacteriophage phi29.FEMS Microbiol Rev. 2010 Sep;34(5):828-41. doi: 10.1111/j.1574-6976.2010.00219.x. Epub 2010 Mar 17. FEMS Microbiol Rev. 2010. PMID: 20412311 Review.
-
Closely related and yet special - how SPβ family phages control lysis-lysogeny decisions.Trends Microbiol. 2025 Apr;33(4):387-396. doi: 10.1016/j.tim.2024.11.007. Epub 2024 Dec 6. Trends Microbiol. 2025. PMID: 39645480 Review.
Cited by
-
Complete genome sequence analysis, morphology and structural protein identification of two Bacillus subtilis phages, BSTP4 and BSTP6, which may form a new species in the genus Salasvirus.Virus Genes. 2023 Aug;59(4):624-634. doi: 10.1007/s11262-023-01998-w. Epub 2023 Apr 29. Virus Genes. 2023. PMID: 37119398
References
-
- Hoch JA. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 1993;47:441–465. - PubMed
-
- Piggot PJ, Losick R. Sporulation genes and intercompartmental regulation. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus Subtilis and its Closest Relatives: From Genes to Cells. Washington, DC: American Society for Microbiology press; 2002. pp. 483–517.
-
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 2003;1:117–126. - PubMed
-
- Lewis RJ, Scott DJ, Brannigan JA, Ladds JC, Cervin MA, Spiegelman GB, Hogget JG, Barák I, Wilkinson AJ. Dimer formation and transcription activation in the sporulation response regulator Spo0A. J. Mol. Biol. 2002;316:235–245. - PubMed
-
- Muchová K, Lewis RJ, Perecko D, Brannigan JA, Ladds JC, Leech A, Wilkinson AJ, Barák I. Dimer-induced signal propagation in Spo0A. Mol. Microbiol. 2004;53:829–842. - PubMed

