The Ime2 protein kinase enhances the disassociation of the Sum1 repressor from middle meiotic promoters
- PMID: 19528232
- PMCID: PMC2725727
- DOI: 10.1128/MCB.00305-09
The Ime2 protein kinase enhances the disassociation of the Sum1 repressor from middle meiotic promoters
Abstract
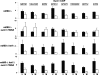
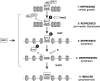
Meiotic development in Saccharomyces cerevisiae (sporulation) is controlled by the sequential transcription of temporally distinct sets of meiosis-specific genes. The induction of middle genes controls exit from meiotic prophase, the completion of the nuclear divisions, and spore formation. Middle promoters are controlled through DNA elements termed middle sporulation elements (MSEs) that are bound by the Sum1 repressor during vegetative growth and by the Ndt80 activator during meiosis. It has been proposed that the induction of middle promoters is controlled by competition between Ndt80 and Sum1 for MSE occupancy. Here, we show that the Sum1 repressor can be removed from middle promoters in meiotic cells independent of Ndt80 expression. This process requires the phosphorylation of Sum1 by the meiosis-specific cyclin-dependent kinase-like kinase Ime2. The deletion of HST1, which encodes a Sir2 paralog that interacts with Sum1, bypasses the requirement for this phosphorylation. These findings suggest that in the presence of Ndt80, Sum1 may be displaced from MSEs through a competition-based mechanism but that in the absence of Ndt80, Sum1 is removed from chromatin in a separate pathway requiring the phosphorylation of Sum1 by Ime2 and the inhibition of Hst1.
Figures


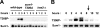
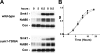
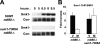
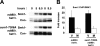

References
-
- Allers, T., and M. Lichten. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 10647-57. - PubMed
-
- Byers, B. 1981. Cytology of the yeast life cycle, p. 59-96. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282699-705. (Erratum, 282:1421.) - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
