The rate of cell growth is governed by cell cycle stage
- PMID: 19528319
- PMCID: PMC2701574
- DOI: 10.1101/gad.1777309
The rate of cell growth is governed by cell cycle stage
Abstract
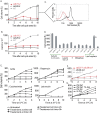
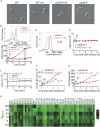
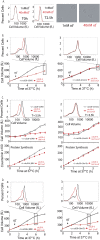
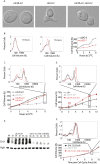
Cell growth is an essential requirement for cell cycle progression. While it is often held that growth is independent of cell cycle position, this relationship has not been closely scrutinized. Here we show that in budding yeast, the ability of cells to grow changes during the cell cycle. We find that cell growth is faster in cells arrested in anaphase and G1 than in other cell cycle stages. We demonstrate that the establishment of a polarized actin cytoskeleton-either as a consequence of normal cell division or through activation of the mating pheromone response-potently attenuates protein synthesis and growth. We furthermore show by population and single-cell analysis that growth varies during an unperturbed cell cycle, slowing at the time of polarized growth. Our study uncovers a fundamental relationship whereby cell cycle position regulates growth.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
