Genome engineering-based analysis of Bearded family genes reveals both functional redundancy and a nonessential function in lateral inhibition in Drosophila
- PMID: 19528324
- PMCID: PMC2728851
- DOI: 10.1534/genetics.109.105023
Genome engineering-based analysis of Bearded family genes reveals both functional redundancy and a nonessential function in lateral inhibition in Drosophila
Abstract
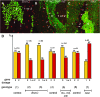
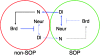
Lateral inhibition mediated by Notch receptor signaling regulates the determination of sensory organ precursor cells (SOPs) in Drosophila. The selection of SOPs from proneural cluster cells appears to rely on a negative feedback loop linking activation of the Notch receptor to downregulation of its ligand Delta within each cell. The molecular basis of this regulatory feedback mechanism is not known. Here, we have tested the role of the Bearded (Brd) family genes in this process. The Drosophila genome encodes eight Brd family members that interact with the E3 ubiquitin ligase Neuralized (Neur) and act as inhibitors of Neur-mediated Delta signaling. Genome engineering technologies were used to create specific deletions of all eight Brd family genes. We find that the Brd family genes malpha, m4, and m6 encoded by the Enhancer of split Complex (E(spl)-C) are dispensable for Drosophila development and that deletion of the five Brd family genes encoded by the Brd Complex only reduces viability. However, deletion of all Brd family genes results in embryonic lethality. Additionally, the malpha, m4, and m6 genes act redundantly with the other five Brd family genes to spatially restrict Notch activation in stage 5 embryos. These data reveal that the Brd family genes have an essential but redundant activity. While the activity of all eight Brd genes appears to be dispensable for SOP determination, clone border studies indicate that both the relative activity levels of Neur and Brd family members influence competition for the SOP fate during lateral inhibition. We propose that inhibition of Neur-Delta interaction by Brd family members is part of the feedback loop that underlies lateral inhibition in Drosophila.
Figures
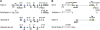


References
-
- Apidianakis, Y., A. C. Nagel, A. Chalkiadaki, A. Preiss and C. Delidakis, 1999. Overexpression of the m4 and malpha genes of the E(spl)-complex antagonizes notch mediated lateral inhibition. Mech. Dev. 86 39–50. - PubMed
-
- Bang, A. G., A. M. Bailey and J. W. Posakony, 1995. Hairless promotes stable commitment to the sensory organ precursor cell fate by negatively regulating the activity of the Notch signaling pathway. Dev. Biol. 172 479–494. - PubMed
-
- Bardin, A. J., and F. Schweisguth, 2006. Bearded family members inhibit Neuralized-mediated endocytosis and signaling activity of Delta in Drosophila. Dev. Cell 10 245–255. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

