In the light of directed evolution: pathways of adaptive protein evolution
- PMID: 19528653
- PMCID: PMC2702793
- DOI: 10.1073/pnas.0901522106
In the light of directed evolution: pathways of adaptive protein evolution
Abstract
Directed evolution is a widely-used engineering strategy for improving the stabilities or biochemical functions of proteins by repeated rounds of mutation and selection. These experiments offer empirical lessons about how proteins evolve in the face of clearly-defined laboratory selection pressures. Directed evolution has revealed that single amino acid mutations can enhance properties such as catalytic activity or stability and that adaptation can often occur through pathways consisting of sequential beneficial mutations. When there are no single mutations that improve a particular protein property experiments always find a wealth of mutations that are neutral with respect to the laboratory-defined measure of fitness. These neutral mutations can open new adaptive pathways by at least 2 different mechanisms. Functionally-neutral mutations can enhance a protein's stability, thereby increasing its tolerance for subsequent functionally beneficial but destabilizing mutations. They can also lead to changes in "promiscuous" functions that are not currently under selective pressure, but can subsequently become the starting points for the adaptive evolution of new functions. These lessons about the coupling between adaptive and neutral protein evolution in the laboratory offer insight into the evolution of proteins in nature.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

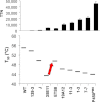

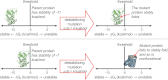
References
-
- Zuckerkandl E, Pauling L. In: Evolutionary Divergence and Convergence in Proteins. Bryson V, Vogel HJ, editors. New York: Springer; 1965. pp. 97–166.
-
- Pauling L, et al. Sickle cell anemia a molecular disease. Science. 1949;110:543–548. - PubMed
-
- Ingram VM. Gene mutations in human hemoglobin: The chemical difference between normal and sickle cell hemoglobin. Nature. 1957;180:326–328. - PubMed
-
- Gillespie JH. The status of the neutral theory: The neutral theory of molecular evolution. Science. 1984;224:732–733. - PubMed
-
- Blum D. Scientists in open war over “neutral theory” of genetics. Sacramento Bee. 1992 Mar 16;:A1.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

