Heterochromatic siRNAs and DDM1 independently silence aberrant 5S rDNA transcripts in Arabidopsis
- PMID: 19529764
- PMCID: PMC2691480
- DOI: 10.1371/journal.pone.0005932
Heterochromatic siRNAs and DDM1 independently silence aberrant 5S rDNA transcripts in Arabidopsis
Abstract
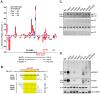
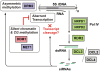
5S ribosomal RNA gene repeats are arranged in heterochromatic arrays (5S rDNA) situated near the centromeres of Arabidopsis chromosomes. The chromatin remodeling factor DDM1 is known to maintain 5S rDNA methylation patterns while silencing transcription through 5S rDNA intergenic spacers (IGS). We mapped small-interfering RNAs (siRNA) to a composite 5S rDNA repeat, revealing a high density of siRNAs matching silenced IGS transcripts. IGS transcript repression requires proteins of the heterochromatic siRNA pathway, including RNA polymerase IV (Pol IV), RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) and DICER-LIKE 3 (DCL3). Using molecular and cytogenetic approaches, we show that the DDM1 and siRNA-dependent silencing effects are genetically independent. DDM1 suppresses production of the siRNAs, however, thereby limiting RNA-directed DNA methylation at 5S rDNA repeats. We conclude that DDM1 and siRNA-dependent silencing are overlapping processes that both repress aberrant 5S rDNA transcription and contribute to the heterochromatic state of 5S rDNA arrays.
Conflict of interest statement
Figures

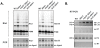


References
-
- Parker JS, Barford D. Argonaute: A scaffold for the function of short regulatory RNAs. Trends Biochem Sci. 2006;31:622–630. - PubMed
-
- Meins F, Jr, Si-Ammour A, Blevins T. RNA silencing systems and their relevance to plant development. Annu Rev Cell Dev Biol. 2005;21:297–318. - PubMed
-
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. - PubMed
-
- Pikaard CS. Cell Biology of the Arabidopsis Nuclear siRNA Pathway for RNA-directed Chromatin Modification. Cold Spring Harb Symp Quant Biol. 2006;71:473–480. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

