A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks
- PMID: 19529817
- PMCID: PMC2639728
- DOI: 10.1093/mp/ssn086
A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks
Erratum in
- Mol Plant. 2015 Mar;8(3):493
Abstract
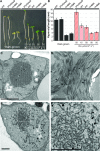
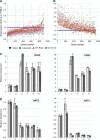
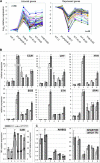
Dominant gain-of-function alleles of Arabidopsis phytochrome B were recently shown to confer light-independent, constitutive photomorphogenic (cop) phenotypes to transgenic plants (Su and Lagarias, 2007). In the present study, comparative transcription profiling experiments were performed to assess whether the pattern of gene expression regulated by these alleles accurately reflects the process of photomorphogenesis in wild-type Arabidopsis. Whole-genome transcription profiles of dark-grown phyAphyB seedlings expressing the Y276H mutant of phyB (YHB) revealed that YHB reprograms about 13% of the Arabidopsis transcriptome in a light-independent manner. The YHB-regulated transcriptome proved qualitatively similar to but quantitatively greater than those of wild-type seedlings grown under 15 or 50 micromol m(-2) m(-1) continuous red light (Rc). Among the 2977 genes statistically significant two-fold (SSTF) regulated by YHB in the absence of light include those encoding components of the photosynthetic apparatus, tetrapyrrole/pigment biosynthetic pathways, and early light-responsive signaling factors. Approximately 80% of genes SSTF regulated by Rc were also YHB-regulated. Expression of a notable subset of 346 YHB-regulated genes proved to be strongly attenuated by Rc, indicating compensating regulation by phyC-E and/or other Rc-dependent processes. Since the majority of these 346 genes are regulated by the circadian clock, these results suggest that phyA- and phyB-independent light signaling pathway(s) strongly influence clock output. Together with the unique plastid morphology of dark-grown YHB seedlings, these analyses indicate that the YHB mutant induces constitutive photomorphogenesis via faithful reconstruction of phyB signaling pathways in a light-independent fashion.
Keywords: Arabidopsis; light signaling; photomorphogenesis; phytochrome; signal transduction; transcriptome analysis.
Figures

References
-
- Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell. 2006;23:439–446. - PubMed
-
- Alabadi D, Yanovsky MJ, Mas P, Harmer SL, Kay SA. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis . Curr. Biol. 2002;12:757–761. - PubMed
-
- Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Ann. Rev. Plant Biol. 2008;59:281–311. - PubMed
-
- Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Ann. Rev. Genet. 2004;38:87–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

