Transcripts of unknown function in multiple-signaling pathways involved in human stem cell differentiation
- PMID: 19531736
- PMCID: PMC2731886
- DOI: 10.1093/nar/gkp426
Transcripts of unknown function in multiple-signaling pathways involved in human stem cell differentiation
Abstract
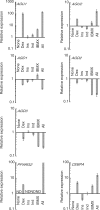
Mammalian transcriptome analysis has uncovered tens of thousands of novel transcripts of unknown function (TUFs). Classical and recent examples suggest that the majority of TUFs may underlie vital intracellular functions as non-coding RNAs because of their low coding potentials. However, only a portion of TUFs have been studied to date, and the functional significance of TUFs remains mostly uncharacterized. To increase the repertoire of functional TUFs, we screened for TUFs whose expression is controlled during differentiation of pluripotent human mesenchymal stem cells (hMSCs). The resulting six TUFs, named transcripts related to hMSC differentiation (TMDs), displayed distinct transcriptional kinetics during hMSC adipogenesis and/or osteogenesis. Structural and comparative genomic characterization suggested a wide variety of biologically active structures of these TMDs, including a long nuclear non-coding RNA, a microRNA host gene and a novel small protein gene. Moreover, the transcriptional response to established pathway activators indicated that most of these TMDs were transcriptionally regulated by each of the two key pathways for hMSC differentiation: the Wnt and protein kinase A (PKA) signaling pathways. The present study suggests that not only TMDs but also other human TUFs may in general participate in vital cellular functions with different molecular mechanisms.
Figures




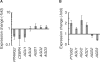
References
-
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. - PubMed
-
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. - PubMed
-
- Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. - PubMed
-
- Kapranov P., Willingham A.T., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. - PubMed
-
- Willingham A.T., Gingeras T.R. TUF love for ‘junk’ DNA. Cell. 2006;125:1215–1220. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

