Host cell transcriptional profiling during malaria liver stage infection reveals a coordinated and sequential set of biological events
- PMID: 19534804
- PMCID: PMC2706893
- DOI: 10.1186/1471-2164-10-270
Host cell transcriptional profiling during malaria liver stage infection reveals a coordinated and sequential set of biological events
Abstract
Background: Plasmodium sporozoites migrate to the liver where they traverse several hepatocytes before invading the one inside which they will develop and multiply into thousands of merozoites. Although this constitutes an essential step of malaria infection, the requirements of Plasmodium parasites in liver cells and how they use the host cell for their own survival and development are poorly understood.
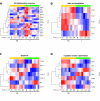
Results: To gain new insights into the molecular host-parasite interactions that take place during malaria liver infection, we have used high-throughput microarray technology to determine the transcriptional profile of P. berghei-infected hepatoma cells. The data analysis shows differential expression patterns for 1064 host genes starting at 6 h and up to 24 h post infection, with the largest proportion correlating specifically with the early stages of the infection process. A considerable proportion of those genes were also found to be modulated in liver cells collected from P. yoelii-infected mice 24 and 40 h after infection, strengthening the data obtained with the in vitro model and highlighting genes and pathways involved in the host response to rodent Plasmodium parasites.
Conclusion: Our data reveal that host cell infection by Plasmodium sporozoites leads to a coordinated and sequential set of biological events, ranging from the initial stage of stress response up to the engagement of host metabolic processes and the maintenance of cell viability throughout infection.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

