Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1
- PMID: 19535331
- PMCID: PMC2755947
- DOI: 10.1074/jbc.M109.029850
Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1
Abstract
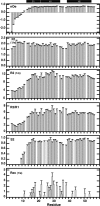
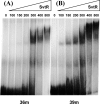
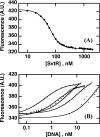
We have characterized the structure and the function of the 6.6-kDa protein SvtR (formerly called gp08) from the rod-shaped virus SIRV1, which infects the hyperthermophilic archaeon Sulfolobus islandicus that thrives at 85 degrees C in hot acidic springs. The protein forms a dimer in solution. The NMR solution structure of the protein consists of a ribbon-helix-helix (RHH) fold between residues 13 and 56 and a disordered N-terminal region (residues 1-12). The structure is very similar to that of bacterial RHH proteins despite the low sequence similarity. We demonstrated that the protein binds DNA and uses its beta-sheet face for the interaction like bacterial RHH proteins. To detect all the binding sites on the 32.3-kb SIRV1 linear genome, we designed and performed a global genome-wide search of targets based on a simplified electrophoretic mobility shift assay. Four targets were recognized by the protein. The strongest binding was observed with the promoter of the gene coding for a virion structural protein. When assayed in a host reconstituted in vitro transcription system, the protein SvtR (Sulfolobus virus transcription regulator) repressed transcription from the latter promoter, as well as from the promoter of its own gene.
Figures
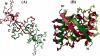


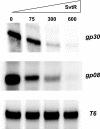
Similar articles
-
Transcription of the rod-shaped viruses SIRV1 and SIRV2 of the hyperthermophilic archaeon sulfolobus.J Bacteriol. 2004 Nov;186(22):7745-53. doi: 10.1128/JB.186.22.7745-7753.2004. J Bacteriol. 2004. PMID: 15516589 Free PMC article.
-
A novel archaeal regulatory protein, Sta1, activates transcription from viral promoters.Nucleic Acids Res. 2006;34(17):4837-45. doi: 10.1093/nar/gkl502. Epub 2006 Sep 14. Nucleic Acids Res. 2006. PMID: 16973899 Free PMC article.
-
Protein-protein interactions leading to recruitment of the host DNA sliding clamp by the hyperthermophilic Sulfolobus islandicus rod-shaped virus 2.J Virol. 2014 Jun;88(12):7105-8. doi: 10.1128/JVI.00636-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696494 Free PMC article.
-
Host and viral transcriptional regulators in Sulfolobus: an overview.Extremophiles. 2013 Nov;17(6):881-95. doi: 10.1007/s00792-013-0586-9. Epub 2013 Oct 2. Extremophiles. 2013. PMID: 24085522 Review.
-
Genomics and biology of Rudiviruses, a model for the study of virus-host interactions in Archaea.Biochem Soc Trans. 2013 Feb 1;41(1):443-50. doi: 10.1042/BST20120313. Biochem Soc Trans. 2013. PMID: 23356326 Free PMC article. Review.
Cited by
-
Solution structure of an archaeal DNA binding protein with an eukaryotic zinc finger fold.PLoS One. 2013;8(1):e52908. doi: 10.1371/journal.pone.0052908. Epub 2013 Jan 9. PLoS One. 2013. PMID: 23326363 Free PMC article.
-
Differentiation and Structure in Sulfolobus islandicus Rod-Shaped Virus Populations.Viruses. 2017 May 19;9(5):120. doi: 10.3390/v9050120. Viruses. 2017. PMID: 28534836 Free PMC article.
-
A conserved hexanucleotide motif is important in UV-inducible promoters in Sulfolobus acidocaldarius.Microbiology (Reading). 2017 May;163(5):778-788. doi: 10.1099/mic.0.000455. Epub 2017 May 3. Microbiology (Reading). 2017. PMID: 28463103 Free PMC article.
-
Archaeal extrachromosomal genetic elements.Microbiol Mol Biol Rev. 2015 Mar;79(1):117-52. doi: 10.1128/MMBR.00042-14. Microbiol Mol Biol Rev. 2015. PMID: 25694123 Free PMC article. Review.
-
A clade of RHH proteins ubiquitous in Sulfolobales and their viruses regulates cell cycle progression.Nucleic Acids Res. 2023 Feb 28;51(4):1724-1739. doi: 10.1093/nar/gkad011. Nucleic Acids Res. 2023. PMID: 36727447 Free PMC article.
References
-
- Torsvik T., Dundas I. D. (1974) Nature 248, 680–681 - PubMed
-
- Prangishvili D., Stedman K., Zillig W. (2001) Trends Microbiol. 9, 39–43 - PubMed
-
- Prangishvili D., Garrett R. A. (2004) Biochem. Soc. Trans. 32, 204–208 - PubMed
-
- Prangishvili D., Forterre P., Garrett R. A. (2006) Nat. Rev. Microbiol. 4, 837–848 - PubMed
-
- Prangishvili D., Garrett R. A., Koonin E. V. (2006) Virus Res. 117, 52–67 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

