Printor, a novel torsinA-interacting protein implicated in dystonia pathogenesis
- PMID: 19535332
- PMCID: PMC2755898
- DOI: 10.1074/jbc.M109.004838
Printor, a novel torsinA-interacting protein implicated in dystonia pathogenesis
Abstract
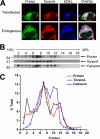
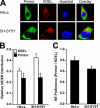
Early onset generalized dystonia (DYT1) is an autosomal dominant neurological disorder caused by deletion of a single glutamate residue (torsinA DeltaE) in the C-terminal region of the AAA(+) (ATPases associated with a variety of cellular activities) protein torsinA. The pathogenic mechanism by which torsinA DeltaE mutation leads to dystonia remains unknown. Here we report the identification and characterization of a 628-amino acid novel protein, printor, that interacts with torsinA. Printor co-distributes with torsinA in multiple brain regions and co-localizes with torsinA in the endoplasmic reticulum. Interestingly, printor selectively binds to the ATP-free form but not to the ATP-bound form of torsinA, supporting a role for printor as a cofactor rather than a substrate of torsinA. The interaction of printor with torsinA is completely abolished by the dystonia-associated torsinA DeltaE mutation. Our findings suggest that printor is a new component of the DYT1 pathogenic pathway and provide a potential molecular target for therapeutic intervention in dystonia.
Figures



References
-
- Bressman S. B., Fahn S., Ozelius L. J., Kramer P. L., Risch N. J. (2001) Arch. Neurol. 58, 681–682 - PubMed
-
- Walker R. H., Brin M. F., Sandu D., Good P. F., Shashidharan P. (2002) Neurology 58, 120–124 - PubMed
-
- Rostasy K., Augood S. J., Hewett J. W., Leung J. C., Sasaki H., Ozelius L. J., Ramesh V., Standaert D. G., Breakefield X. O., Hedreen J. C. (2003) Neurobiol. Dis. 12, 11–24 - PubMed
-
- Ozelius L. J., Hewett J. W., Page C. E., Bressman S. B., Kramer P. L., Shalish C., de Leon D., Brin M. F., Raymond D., Corey D. P., Fahn S., Risch N. J., Buckler A. J., Gusella J. F., Breakefield X. O. (1997) Nat. Genet. 17, 40–48 - PubMed
-
- Leung J. C., Klein C., Friedman J., Vieregge P., Jacobs H., Doheny D., Kamm C., DeLeon D., Pramstaller P. P., Penney J. B., Eisengart M., Jankovic J., Gasser T., Bressman S. B., Corey D. P., Kramer P., Brin M. F., Ozelius L. J., Breakefield X. O. (2001) Neurogenetics 3, 133–143 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

