Comparative prion disease gene expression profiling using the prion disease mimetic, cuprizone
- PMID: 19535908
- PMCID: PMC2712607
- DOI: 10.4161/pri.3.2.9059
Comparative prion disease gene expression profiling using the prion disease mimetic, cuprizone
Abstract
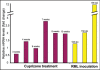
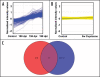
Identification of genes expressed in response to prion infection may elucidate biomarkers for disease, identify factors involved in agent replication, mechanisms of neuropathology and therapeutic targets. Although several groups have sought to identify gene expression changes specific to prion disease, expression profiles rife with cell population changes have consistently been identified. Cuprizone, a neurotoxicant, qualitatively mimics the cell population changes observed in prion disease, resulting in both spongiform change and astrocytosis. The use of cuprizone-treated animals as an experimental control during comparative expression profiling allows for the identification of transcripts whose expression increases during prion disease and remains unchanged during cuprizone-triggered neuropathology. In this study, expression profiles from the brains of mice preclinically and clinically infected with Rocky Mountain Laboratory (RML) mouse-adapted scrapie agent and age-matched controls were profiled using Affymetrix gene arrays. In total, 164 genes were differentially regulated during prion infection. Eighty-three of these transcripts have been previously undescribed as differentially regulated during prion disease. A 0.4% cuprizone diet was utilized as a control for comparative expression profiling. Cuprizone treatment induced spongiosis and astrocyte proliferation as indicated by glial fibrillary acidic protein (Gfap) transcriptional activation and immunohistochemistry. Gene expression profiles from brain tissue obtained from cuprizone-treated mice identified 307 differentially regulated transcript changes. After comparative analysis, 17 transcripts unaffected by cuprizone treatment but increasing in expression from preclinical to clinical prion infection were identified. Here we describe the novel use of the prion disease mimetic, cuprizone, to control for cell population changes in the brain during prion infection.
Figures




Similar articles
-
Molecular signatures in prion disease: altered death receptor pathways in a mouse model.J Transl Med. 2024 May 27;22(1):503. doi: 10.1186/s12967-024-05121-x. J Transl Med. 2024. PMID: 38802941 Free PMC article.
-
Identification of differentially expressed genes in scrapie-infected mouse brains by using global gene expression technology.J Virol. 2004 Oct;78(20):11051-60. doi: 10.1128/JVI.78.20.11051-11060.2004. J Virol. 2004. PMID: 15452225 Free PMC article.
-
RNA-seq and network analysis reveal unique glial gene expression signatures during prion infection.Mol Brain. 2020 May 7;13(1):71. doi: 10.1186/s13041-020-00610-8. Mol Brain. 2020. PMID: 32381108 Free PMC article.
-
Prion protein transgenes and the neuropathology in prion diseases.Brain Pathol. 1995 Jan;5(1):77-89. doi: 10.1111/j.1750-3639.1995.tb00579.x. Brain Pathol. 1995. PMID: 7767493 Review.
-
The neuropathological phenotype in transgenic mice expressing different prion protein constructs.Philos Trans R Soc Lond B Biol Sci. 1994 Mar 29;343(1306):415-23. doi: 10.1098/rstb.1994.0038. Philos Trans R Soc Lond B Biol Sci. 1994. PMID: 7913760 Review.
Cited by
-
Regional Differences in Neuroinflammation-Associated Gene Expression in the Brain of Sporadic Creutzfeldt-Jakob Disease Patients.Int J Mol Sci. 2020 Dec 25;22(1):140. doi: 10.3390/ijms22010140. Int J Mol Sci. 2020. PMID: 33375642 Free PMC article.
-
Down-regulation of Shadoo in prion infections traces a pre-clinical event inversely related to PrP(Sc) accumulation.PLoS Pathog. 2011 Nov;7(11):e1002391. doi: 10.1371/journal.ppat.1002391. Epub 2011 Nov 17. PLoS Pathog. 2011. PMID: 22114562 Free PMC article.
-
Upregulation of interferon-gamma-induced genes during prion infection.J Toxicol Environ Health A. 2011;74(2-4):146-53. doi: 10.1080/15287394.2011.529064. J Toxicol Environ Health A. 2011. PMID: 21218343 Free PMC article.
-
Toll-like receptor 2 confers partial neuroprotection during prion disease.PLoS One. 2018 Dec 31;13(12):e0208559. doi: 10.1371/journal.pone.0208559. eCollection 2018. PLoS One. 2018. PMID: 30596651 Free PMC article.
-
Transcriptomic responses to prion disease in rats.BMC Genomics. 2015 Sep 5;16(1):682. doi: 10.1186/s12864-015-1884-7. BMC Genomics. 2015. PMID: 26341492 Free PMC article.
References
-
- Collinge J. Prion diseases of humans and animals: Their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
