Cep72 regulates the localization of key centrosomal proteins and proper bipolar spindle formation
- PMID: 19536135
- PMCID: PMC2718283
- DOI: 10.1038/emboj.2009.161
Cep72 regulates the localization of key centrosomal proteins and proper bipolar spindle formation
Abstract
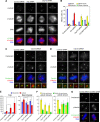
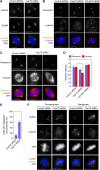
Microtubule-nucleation activity and structural integrity of the centrosome are critical for various cellular functions. The gamma-tubulin ring complexes (gammaTuRCs) localizing to the pericentriolar matrix (PCM) of the centrosome are major sites of microtubule nucleation. The PCM is thought to be created by two cognate large coiled-coil proteins, pericentrin/kendrin and CG-NAP/AKAP450, and its stabilization by Kizuna is essential for bipolar spindle formation. However, the mechanisms by which these proteins are recruited and organized into a proper structure with microtubule-organizing activity are poorly understood. Here we identify a centrosomal protein Cep72 as a Kizuna-interacting protein. Interestingly, Cep72 is essential for the localization of CG-NAP and Kizuna. Cep72 is also involved in gammaTuRC recruitment to the centrosome and CG-NAP confers the microtubule-nucleation activity on the gammaTuRCs. During mitosis, Cep72-mediated microtubule organization is important for converging spindle microtubules to the centrosomes, which is needed for chromosome alignment and tension generation between kinetochores. Our findings show that Cep72 is the key protein essential for maintaining microtubule-organizing activity and structural integrity of the centrosome.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 - PubMed
-
- Bettencourt-Dias M, Glover DM (2007) Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol 8: 451–463 - PubMed
-
- Blagden SP, Glover DM (2003) Polar expeditions—provisioning the centrosome for mitosis. Nat Cell Biol 5: 505–511 - PubMed
-
- Bornens M (2002) Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol 14: 25–34 - PubMed
-
- Casenghi M, Meraldi P, Weinhart U, Duncan PI, Korner R, Nigg EA (2003) Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev Cell 5: 113–125 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

