Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps
- PMID: 19536157
- PMCID: PMC2733940
- DOI: 10.1038/nature08170
Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps
Abstract
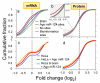
MicroRNAs (miRNAs) have critical roles in the regulation of gene expression; however, as miRNA activity requires base pairing with only 6-8 nucleotides of messenger RNA, predicting target mRNAs is a major challenge. Recently, high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (HITS-CLIP) has identified functional protein-RNA interaction sites. Here we use HITS-CLIP to covalently crosslink native argonaute (Ago, also called Eif2c) protein-RNA complexes in mouse brain. This produced two simultaneous data sets-Ago-miRNA and Ago-mRNA binding sites-that were combined with bioinformatic analysis to identify interaction sites between miRNA and target mRNA. We validated genome-wide interaction maps for miR-124, and generated additional maps for the 20 most abundant miRNAs present in P13 mouse brain. Ago HITS-CLIP provides a general platform for exploring the specificity and range of miRNA action in vivo, and identifies precise sequences for targeting clinically relevant miRNA-mRNA interactions.
Figures

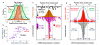


References
-
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. - PubMed
-
- Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. - PubMed
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

