Prion protein paralog doppel protein interacts with alpha-2-macroglobulin: a plausible mechanism for doppel-mediated neurodegeneration
- PMID: 19536284
- PMCID: PMC2693666
- DOI: 10.1371/journal.pone.0005968
Prion protein paralog doppel protein interacts with alpha-2-macroglobulin: a plausible mechanism for doppel-mediated neurodegeneration
Erratum in
- PLoS One. 2009;4(8). doi: 10.1371/annotation/6f581801-c5f0-4fb0-9e55-6ee35653cd12
Abstract
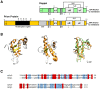
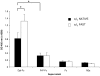
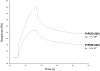
Doppel protein (Dpl) is a paralog of the cellular form of the prion protein (PrP(C)), together sharing common structural and biochemical properties. Unlike PrP(C), which is abundantly expressed throughout the central nervous system (CNS), Dpl protein expression is not detectable in the CNS. Interestingly, its ectopic expression in the brain elicits neurodegeneration in transgenic mice. Here, by combining native isoelectric focusing plus non-denaturing polyacrylamide gel electrophoresis and mass spectrometry analysis, we identified two Dpl binding partners: rat alpha-1-inhibitor-3 (alpha(1)I(3)) and, by sequence homology, alpha-2-macroglobulin (alpha(2)M), two known plasma metalloproteinase inhibitors. Biochemical investigations excluded the direct interaction of PrP(C) with either alpha(1)I(3) or alpha(2)M. Nevertheless, enzyme-linked immunosorbent assays and surface plasmon resonance experiments revealed a high affinity binding occurring between PrP(C) and Dpl. In light of these findings, we suggest a mechanism for Dpl-induced neurodegeneration in mice expressing Dpl ectopically in the brain, linked to a withdrawal of natural inhibitors of metalloproteinase such as alpha(2)M. Interestingly, alpha(2)M has been proven to be a susceptibility factor in Alzheimer's disease, and as our findings imply, it may also play a relevant role in other neurodegenerative disorders, including prion diseases.
Conflict of interest statement
Figures






References
-
- Prusiner SB. Prions (Les Prix Nobel Lecture). In: Frängsmyr T, editor. Les Prix Nobel. Stockholm, Sweden: Almqvist & Wiksell International; 1998. pp. 268–323.
-
- Prusiner SB. Shattuck Lecture — Neurodegenerative diseases and prions. N Engl J Med. 2001;344:1516–1526. - PubMed
-
- Moore RC, Lee IY, Silverman GL, Harrison PM, Strome R, et al. Ataxia in prion protein (PrP)-deficient mice is associated with upregulation of the novel PrP-like protein doppel. J Mol Biol. 1999;292:797–817. - PubMed
-
- Silverman GL, Qin K, Moore RC, Yang Y, Mastrangelo P, et al. Doppel is an N-glycosylated, glycosylphosphatidylinositol-anchored protein. Expression in testis and ectopic production in the brains of Prnp(0/0) mice predisposed to Purkinje cell loss. J Biol Chem. 2000;275:26834–26841. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

