The complete genome of klassevirus - a novel picornavirus in pediatric stool
- PMID: 19538752
- PMCID: PMC2709156
- DOI: 10.1186/1743-422X-6-82
The complete genome of klassevirus - a novel picornavirus in pediatric stool
Abstract
Background: Diarrhea kills 2 million children worldwide each year, yet an etiological agent is not found in approximately 30-50% of cases. Picornaviral genera such as enterovirus, kobuvirus, cosavirus, parechovirus, hepatovirus, teschovirus, and cardiovirus have all been found in human and animal diarrhea. Modern technologies, especially deep sequencing, allow rapid, high-throughput screening of clinical samples such as stool for new infectious agents associated with human disease.
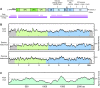
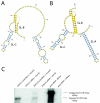
Results: A pool of 141 pediatric gastroenteritis samples that were previously found to be negative for known diarrheal viruses was subjected to pyrosequencing. From a total of 937,935 sequence reads, a collection of 849 reads distantly related to Aichi virus were assembled and found to comprise 75% of a novel picornavirus genome. The complete genome was subsequently cloned and found to share 52.3% nucleotide pairwise identity and 38.9% amino acid identity to Aichi virus. The low level of sequence identity suggests a novel picornavirus genus which we have designated klassevirus. Blinded screening of 751 stool specimens from both symptomatic and asymptomatic individuals revealed a second positive case of klassevirus infection, which was subsequently found to be from the index case's 11-month old twin.
Conclusion: We report the discovery of human klassevirus 1, a member of a novel picornavirus genus, in stool from two infants from Northern California. Further characterization and epidemiological studies will be required to establish whether klasseviruses are significant causes of human infection.
Figures


References
-
- Racaniello VR. Picornaviridae: The Viruses and Their Replication. In: David M Knipe, Peter M, editor. Fields Virology. 5. Vol. 1. Howley: Lippincott Williams & Wilkins; 2007. pp. 795–838.
-
- Yamashita T, Kobayashi S, Sakae K, Nakata S, Chiba S, Ishihara Y, Isomura S. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J Infect Dis. 1991;164:954–7. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

