Instability and chromatin structure of expanded trinucleotide repeats
- PMID: 19540013
- PMCID: PMC3671858
- DOI: 10.1016/j.tig.2009.04.007
Instability and chromatin structure of expanded trinucleotide repeats
Abstract
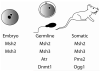
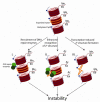
Trinucleotide repeat expansion underlies at least 17 neurological diseases. In affected individuals, the expanded locus is characterized by dramatic changes in chromatin structure and in repeat tract length. Interestingly, recent studies show that several chromatin modifiers, including a histone acetyltransferase, a DNA methyltransferase and the chromatin insulator CTCF can modulate repeat instability. Here, we propose that the unusual chromatin structure of expanded repeats directly impacts their instability. We discuss several potential models for how this might occur, including a role for DNA repair-dependent epigenetic reprogramming in increasing repeat instability, and the capacity of epigenetic marks to alter sense and antisense transcription, thereby affecting repeat instability.
Figures
References
-
- Verkerk AJ, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. - PubMed
-
- La Spada AR, et al. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. - PubMed
-
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. - PubMed
-
- Pearson CE, et al. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. - PubMed
-
- Libby RT, et al. Genomic context drives SCA7 CAG repeat instability, while expressed SCA7 cDNAs are intergenerationally and somatically stable in transgenic mice. Hum Mol Genet. 2003;12:41–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

