Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells
- PMID: 19540188
- PMCID: PMC2720132
- DOI: 10.1016/j.stem.2009.05.023
Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells
Abstract
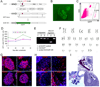
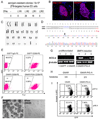
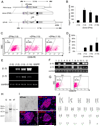
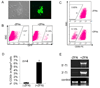
We report here homologous recombination (HR)-mediated gene targeting of two different genes in human iPS cells (hiPSCs) and human ES cells (hESCs). HR-mediated correction of a chromosomally integrated mutant GFP reporter gene reaches efficiencies of 0.14%-0.24% in both cell types transfected by donor DNA with plasmids expressing zinc finger nucleases (ZFNs). Engineered ZFNs that induce a sequence-specific double-strand break in the GFP gene enhanced HR-mediated correction by > 1400-fold without detectable alterations in stem cell karyotypes or pluripotency. Efficient HR-mediated insertional mutagenesis was also achieved at the endogenous PIG-A locus, with a > 200-fold enhancement by ZFNs targeted to that gene. Clonal PIG-A null hESCs and iPSCs with normal karyotypes were readily obtained. The phenotypic and biological defects were rescued by PIG-A transgene expression. Our study provides the first demonstration of HR-mediated gene targeting in hiPSCs and shows the power of ZFNs for inducing specific genetic modifications in hiPSCs, as well as hESCs.
Figures
Similar articles
-
Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases.Nat Biotechnol. 2009 Sep;27(9):851-7. doi: 10.1038/nbt.1562. Epub 2009 Aug 13. Nat Biotechnol. 2009. PMID: 19680244 Free PMC article.
-
Low-Dose Irradiation Enhances Gene Targeting in Human Pluripotent Stem Cells.Stem Cells Transl Med. 2015 Sep;4(9):998-1010. doi: 10.5966/sctm.2015-0050. Epub 2015 Jul 16. Stem Cells Transl Med. 2015. PMID: 26185257 Free PMC article.
-
Design and testing of zinc finger nucleases for use in mammalian cells.Methods Mol Biol. 2008;435:47-61. doi: 10.1007/978-1-59745-232-8_4. Methods Mol Biol. 2008. PMID: 18370067
-
Concise review: putting a finger on stem cell biology: zinc finger nuclease-driven targeted genetic editing in human pluripotent stem cells.Stem Cells. 2011 Jul;29(7):1021-33. doi: 10.1002/stem.658. Stem Cells. 2011. PMID: 21544904 Review.
-
Generation of GFP Reporter Human Induced Pluripotent Stem Cells Using AAVS1 Safe Harbor Transcription Activator-Like Effector Nuclease.Curr Protoc Stem Cell Biol. 2014 May 16;29:5A.7.1-18. doi: 10.1002/9780470151808.sc05a07s29. Curr Protoc Stem Cell Biol. 2014. PMID: 24838915 Free PMC article. Review.
Cited by
-
Pluripotent stem cells in research and treatment of hemoglobinopathies.Cold Spring Harb Perspect Med. 2012 Apr;2(4):a011841. doi: 10.1101/cshperspect.a011841. Cold Spring Harb Perspect Med. 2012. PMID: 22474618 Free PMC article. Review.
-
Opportunities and challenges of pluripotent stem cell neurodegenerative disease models.Nat Neurosci. 2013 Jul;16(7):780-9. doi: 10.1038/nn.3425. Epub 2013 Jun 25. Nat Neurosci. 2013. PMID: 23799470 Review.
-
Gene knockout and knockin by zinc-finger nucleases: current status and perspectives.Cell Mol Life Sci. 2013 Aug;70(16):2969-83. doi: 10.1007/s00018-012-1204-1. Epub 2012 Nov 17. Cell Mol Life Sci. 2013. PMID: 23161061 Free PMC article. Review.
-
Reshaping pluripotent stem cells.Nat Biotechnol. 2009 Sep;27(9):823-4. doi: 10.1038/nbt0909-823. Nat Biotechnol. 2009. PMID: 19741639 No abstract available.
-
Analysis of illegitimate genomic integration mediated by zinc-finger nucleases: implications for specificity of targeted gene correction.BMC Mol Biol. 2010 May 10;11:35. doi: 10.1186/1471-2199-11-35. BMC Mol Biol. 2010. PMID: 20459736 Free PMC article.
References
-
- Cai L, Ye Z, Zhou BY, Mali P, Zhou C, Cheng L. Promoting human embryonic stem cell renewal or differentiation by modulating Wnt signal and culture conditions. Cell Res. 2007;17:62–72. - PubMed
-
- Cathomen T, Joung KJ. Zinc-finger nucleases: the next generation emerges. Mol Ther. 2008;16:1200–1207. - PubMed
-
- Costa M, Dottori M, Sourris K, Jamshidi P, Hatzistavrou T, Davis R, Azzola L, Jackson S, Lim SM, Pera M, et al. A method for genetic modification of human embryonic stem cells using electroporation. Nat Protoc. 2007;2:792–796. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

