The glucanosyltransferase Gas1 functions in transcriptional silencing
- PMID: 19541632
- PMCID: PMC2708717
- DOI: 10.1073/pnas.0900809106
The glucanosyltransferase Gas1 functions in transcriptional silencing
Abstract
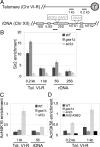
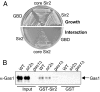
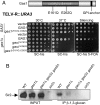
Transcriptional silencing is a crucial process that is mediated through chromatin structure. The histone deacetylase Sir2 silences genomic regions that include telomeres, ribosomal DNA (rDNA) and the cryptic mating-type loci. Here, we report an unsuspected role for the enzyme Gas1 in locus-specific transcriptional silencing. GAS1 encodes a beta-1,3-glucanosyltransferase previously characterized for its role in cell wall biogenesis. In gas1 mutants, telomeric silencing is defective and rDNA silencing is enhanced. We show that the catalytic activity of Gas1 is required for normal silencing, and that Gas1's role in silencing is distinct from its role in cell wall biogenesis. Established hallmarks of silent chromatin, such as Sir2 and Sir3 binding, H4K16 deacetylation, and H3K56 deacetylation, appear unaffected in gas1 mutants. Thus, another event required for telomeric silencing must be influenced by GAS1. Because the catalytic activity of Gas1 is required for telomeric silencing, Gas1 localizes to the nuclear periphery, and Gas1 and Sir2 physically interact, we propose a model in which carbohydrate modification of chromatin components provides a new regulatory element that may be critical for chromatin function but which is virtually unexplored in the current landscape of chromatin analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Fueling transcriptional silencing with Gas1.Proc Natl Acad Sci U S A. 2009 Jul 7;106(27):10879-80. doi: 10.1073/pnas.0905192106. Epub 2009 Jun 30. Proc Natl Acad Sci U S A. 2009. PMID: 19567837 Free PMC article. No abstract available.
Similar articles
-
The β-1,3-glucanosyltransferase Gas1 regulates Sir2-mediated rDNA stability in Saccharomyces cerevisiae.Nucleic Acids Res. 2014 Jul;42(13):8486-99. doi: 10.1093/nar/gku570. Epub 2014 Jun 30. Nucleic Acids Res. 2014. PMID: 24981510 Free PMC article.
-
Fueling transcriptional silencing with Gas1.Proc Natl Acad Sci U S A. 2009 Jul 7;106(27):10879-80. doi: 10.1073/pnas.0905192106. Epub 2009 Jun 30. Proc Natl Acad Sci U S A. 2009. PMID: 19567837 Free PMC article. No abstract available.
-
A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors.Mol Cell Biol. 1999 Apr;19(4):3184-97. doi: 10.1128/MCB.19.4.3184. Mol Cell Biol. 1999. PMID: 10082585 Free PMC article.
-
A model for step-wise assembly of heterochromatin in yeast.Novartis Found Symp. 2004;259:48-56; discussion 56-62, 163-9. Novartis Found Symp. 2004. PMID: 15171246 Review.
-
The Nuts and Bolts of Transcriptionally Silent Chromatin in Saccharomyces cerevisiae.Genetics. 2016 Aug;203(4):1563-99. doi: 10.1534/genetics.112.145243. Genetics. 2016. PMID: 27516616 Free PMC article. Review.
Cited by
-
Unexpected function of the glucanosyltransferase Gas1 in the DNA damage response linked to histone H3 acetyltransferases in Saccharomyces cerevisiae.Genetics. 2014 Apr;196(4):1029-39. doi: 10.1534/genetics.113.158824. Epub 2014 Feb 13. Genetics. 2014. PMID: 24532730 Free PMC article.
-
The C-terminus of histone H2B is involved in chromatin compaction specifically at telomeres, independently of its monoubiquitylation at lysine 123.PLoS One. 2011;6(7):e22209. doi: 10.1371/journal.pone.0022209. Epub 2011 Jul 29. PLoS One. 2011. PMID: 21829450 Free PMC article.
-
The PHR Family: The Role of Extracellular Transglycosylases in Shaping Candida albicans Cells.J Fungi (Basel). 2017 Oct 29;3(4):59. doi: 10.3390/jof3040059. J Fungi (Basel). 2017. PMID: 29371575 Free PMC article. Review.
-
The β-1,3-glucanosyltransferase Gas1 regulates Sir2-mediated rDNA stability in Saccharomyces cerevisiae.Nucleic Acids Res. 2014 Jul;42(13):8486-99. doi: 10.1093/nar/gku570. Epub 2014 Jun 30. Nucleic Acids Res. 2014. PMID: 24981510 Free PMC article.
-
GPI-anchored Gas1 protein regulates cytosolic proteostasis in yeast.bioRxiv [Preprint]. 2023 May 26:2023.05.26.542479. doi: 10.1101/2023.05.26.542479. bioRxiv. 2023. Update in: G3 (Bethesda). 2024 Mar 6;14(3):jkad263. doi: 10.1093/g3journal/jkad263. PMID: 37292646 Free PMC article. Updated. Preprint.
References
-
- Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. - PubMed
-
- Brachmann CB, et al. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. - PubMed
-
- Buck SW, Gallo CM, Smith JS. Diversity in the Sir2 family of protein deacetylases. J Leukoc Biol. 2004;75:939–950. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

