Functional analysis of an Orc6 mutant in Drosophila
- PMID: 19541634
- PMCID: PMC2705598
- DOI: 10.1073/pnas.0902670106
Functional analysis of an Orc6 mutant in Drosophila
Abstract
The origin recognition complex (ORC) is a 6-subunit complex required for the initiation of DNA replication in eukaryotic organisms. ORC is also involved in other cell functions. The smallest Drosophila ORC subunit, Orc6, is important for both DNA replication and cytokinesis. To study the role of Orc6 in vivo, the orc6 gene was deleted by imprecise excision of P element. Lethal alleles of orc6 are defective in DNA replication and also show abnormal chromosome condensation and segregation. The analysis of cells containing the orc6 deletion revealed that they arrest in both the G(1) and mitotic stages of the cell cycle. Orc6 deletion can be rescued to viability by a full-length Orc6 transgene. The expression of mutant transgenes of Orc6 with deleted or mutated C-terminal domain results in a release of mutant cells from G(1) arrest and restoration of DNA replication, indicating that the DNA replication function of Orc6 is associated with its N-terminal domain. However, these mutant cells accumulate at mitosis, suggesting that the C-terminal domain of Orc6 is important for the passage through the M phase. In a cross-species complementation experiment, the expression of human Orc6 in Drosophila Orc6 mutant cells rescued DNA replication, suggesting that this function of the protein is conserved among metazoans.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
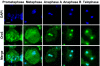

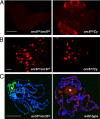



References
-
- Bell SP. The origin recognition complex: From simple origins to complex functions. Genes Dev. 2002;16:659–672. - PubMed
-
- Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: Replication initiation in metazoans. Cell. 2005;123:13–24. - PubMed
-
- Chesnokov IN. Multiple functions of the origin recognition complex. Int Rev Cytol. 2007;256:69–109. - PubMed
-
- Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996;6:1416–1425. - PubMed
-
- Rowles A, et al. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

