Direct observation of an ensemble of stable collapsed states in the mechanical folding of ubiquitin
- PMID: 19541635
- PMCID: PMC2705594
- DOI: 10.1073/pnas.0901213106
Direct observation of an ensemble of stable collapsed states in the mechanical folding of ubiquitin
Abstract
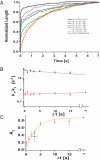
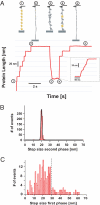
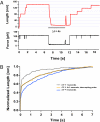
Statistical theories of protein folding have long predicted plausible mechanisms for reducing the vast conformational space through distinct ensembles of structures. However, these predictions have remained untested by bulk techniques, because the conformational diversity of folding molecules has been experimentally unapproachable. Owing to recent advances in single molecule force-clamp spectroscopy, we are now able to probe the structure and dynamics of the small protein ubiquitin by measuring its length and mechanical stability during each stage of folding. Here, we discover that upon hydrophobic collapse, the protein rapidly selects a subset of minimum energy structures that are mechanically weak and essential precursors of the native fold. From this much reduced ensemble, the native state is acquired through a barrier-limited transition. Our results support the validity of statistical mechanics models in describing the folding of a small protein on biological timescales.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Osmolyte-induced separation of the mechanical folding phases of ubiquitin.Proc Natl Acad Sci U S A. 2009 Jun 30;106(26):10540-5. doi: 10.1073/pnas.0902090106. Epub 2009 Jun 16. Proc Natl Acad Sci U S A. 2009. PMID: 19541633 Free PMC article.
-
The folding pathway of ubiquitin from all-atom molecular dynamics simulations.Biophys Chem. 2004 Oct 1;111(2):159-71. doi: 10.1016/j.bpc.2004.05.009. Biophys Chem. 2004. PMID: 15381313
-
Fluctuations of primary ubiquitin folding intermediates in a force clamp.J Struct Biol. 2007 Mar;157(3):557-69. doi: 10.1016/j.jsb.2006.11.005. Epub 2006 Nov 26. J Struct Biol. 2007. PMID: 17306561
-
Conformational dynamics and ensembles in protein folding.Annu Rev Biophys Biomol Struct. 2007;36:395-412. doi: 10.1146/annurev.biophys.36.040306.132608. Annu Rev Biophys Biomol Struct. 2007. PMID: 17291180 Review.
-
Ubiquitin: a small protein folding paradigm.Org Biomol Chem. 2006 May 21;4(10):1845-53. doi: 10.1039/b600829c. Epub 2006 Apr 27. Org Biomol Chem. 2006. PMID: 16688326 Review.
Cited by
-
Probing osmolyte participation in the unfolding transition state of a protein.Proc Natl Acad Sci U S A. 2011 Jun 14;108(24):9759-64. doi: 10.1073/pnas.1101934108. Epub 2011 May 25. Proc Natl Acad Sci U S A. 2011. Retraction in: Proc Natl Acad Sci U S A. 2011 Dec 20;108(51):20850. doi: 10.1073/pnas.1118432108. PMID: 21613570 Free PMC article. Retracted.
-
A simple two-state protein unfolds mechanically via multiple heterogeneous pathways at single-molecule resolution.Nat Commun. 2016 Jun 1;7:11777. doi: 10.1038/ncomms11777. Nat Commun. 2016. PMID: 27248054 Free PMC article.
-
Domain-domain interactions in filamin A (16-23) impose a hierarchy of unfolding forces.Biophys J. 2013 May 7;104(9):2022-30. doi: 10.1016/j.bpj.2013.03.034. Biophys J. 2013. PMID: 23663845 Free PMC article.
-
Full reconstruction of a vectorial protein folding pathway by atomic force microscopy and molecular dynamics simulations.J Biol Chem. 2010 Dec 3;285(49):38167-72. doi: 10.1074/jbc.M110.179697. Epub 2010 Sep 24. J Biol Chem. 2010. PMID: 20870713 Free PMC article.
-
Kinetics of allosteric transitions in S-adenosylmethionine riboswitch are accurately predicted from the folding landscape.J Am Chem Soc. 2013 Nov 6;135(44):16641-50. doi: 10.1021/ja408595e. Epub 2013 Oct 22. J Am Chem Soc. 2013. PMID: 24087850 Free PMC article.
References
-
- Degennes PG. Kinetics of collapse for a flexible coil. J Phys Lett-Paris. 1985;46:L639–L642.
-
- Bryngelson JD, wolynes PG. Intermediates and barrier crossing in random energy model (with applications to protein folding) J Phys Chem. 1989;93:6902–6915.
-
- Bryngelson JD, Wolynes PG. A simple statistical field theory of heteropolymer collpase with application to protein folding. Biopolymers. 1990;30:177–188.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

