Computing protein stabilities from their chain lengths
- PMID: 19541647
- PMCID: PMC2705543
- DOI: 10.1073/pnas.0903995106
Computing protein stabilities from their chain lengths
Abstract
New amino acid sequences of proteins are being learned at a rapid rate, thanks to modern genomics. The native structures and functions of those proteins can often be inferred using bioinformatics methods. We show here that it is also possible to infer the stabilities and thermal folding properties of proteins, given only simple genomics information: the chain length and the numbers of charged side chains. In particular, our model predicts DeltaH(T), DeltaS(T), DeltaC(p), and DeltaF(T)--the folding enthalpy, entropy, heat capacity, and free energy--as functions of temperature T; the denaturant m values in guanidine and urea; the pH-temperature-salt phase diagrams, and the energy of confinement F(s) of the protein inside a cavity of radius s. All combinations of these phase equilibria can also then be computed from that information. As one illustration, we compute the pH and salt conditions that would denature a protein inside a small confined cavity. Because the model is analytical, it is computationally efficient enough that it could be used to automatically annotate whole proteomes with protein stability information.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
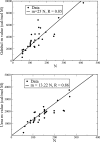



Similar articles
-
Protein stabilization by urea and guanidine hydrochloride.Biochemistry. 2002 Nov 12;41(45):13386-94. doi: 10.1021/bi020371n. Biochemistry. 2002. PMID: 12416983
-
Hydrophobic hydration processes thermal and chemical denaturation of proteins.Biophys Chem. 2011 Jun;156(1):51-67. doi: 10.1016/j.bpc.2011.02.009. Epub 2011 Mar 3. Biophys Chem. 2011. PMID: 21482019
-
Thermodynamics of denaturation of alpha-chymotrypsinogen A in aqueous urea and alkylurea solutions.J Protein Chem. 1995 Nov;14(8):709-19. doi: 10.1007/BF01886910. J Protein Chem. 1995. PMID: 8747432
-
Close-range electrostatic interactions in proteins.Chembiochem. 2002 Jul 2;3(7):604-17. doi: 10.1002/1439-7633(20020703)3:7<604::AID-CBIC604>3.0.CO;2-X. Chembiochem. 2002. PMID: 12324994 Review.
-
Challenges in the computational design of proteins.J R Soc Interface. 2009 Aug 6;6 Suppl 4(Suppl 4):S477-91. doi: 10.1098/rsif.2008.0508.focus. Epub 2009 Mar 11. J R Soc Interface. 2009. PMID: 19324680 Free PMC article. Review.
Cited by
-
Distribution and solvent exposure of Hsp70 chaperone binding sites across the Escherichia coli proteome.Proteins. 2023 May;91(5):665-678. doi: 10.1002/prot.26456. Epub 2023 Jan 1. Proteins. 2023. PMID: 36539330 Free PMC article.
-
A Thermodynamic Atlas of Proteomes Reveals Energetic Innovation across the Tree of Life.Mol Biol Evol. 2022 Mar 2;39(3):msac010. doi: 10.1093/molbev/msac010. Mol Biol Evol. 2022. PMID: 35038744 Free PMC article.
-
The variable domain from dynamin-related protein 1 promotes liquid-liquid phase separation that enhances its interaction with cardiolipin-containing membranes.Protein Sci. 2023 Nov;32(11):e4787. doi: 10.1002/pro.4787. Protein Sci. 2023. PMID: 37743569 Free PMC article.
-
Highly Abundant Proteins Are Highly Thermostable.Genome Biol Evol. 2023 Jul 3;15(7):evad112. doi: 10.1093/gbe/evad112. Genome Biol Evol. 2023. PMID: 37399326 Free PMC article.
-
The Effects of Chain Length on the Structural Properties of Intrinsically Disordered Proteins in Concentrated Solutions.J Phys Chem B. 2020 Dec 31;124(52):11843-11853. doi: 10.1021/acs.jpcb.0c09635. Epub 2020 Dec 18. J Phys Chem B. 2020. PMID: 33337879 Free PMC article.
References
-
- Chen JM, et al. Packing is a key selection factor in the evolution of protein hydrophobic cores. Biochemistry. 2001;40:14004–14011. - PubMed
-
- Plaxco KW, et al. Topology, stability, sequence, and length: Defining the determinants of two-state protein folding kinetics. Biochemistry. 2000;39:11177–11183. - PubMed
-
- Privalov PL, et al. Thermodynamic approach to problem of stabilization of globular protein structure—calorimetric study. J Mol Biol. 1974;86:665–684. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

