Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing
- PMID: 19541912
- PMCID: PMC2765271
- DOI: 10.1101/gr.092924.109
Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing
Abstract
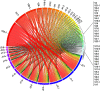
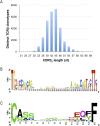
T-cell receptor (TCR) genomic loci undergo somatic V(D)J recombination, plus the addition/subtraction of nontemplated bases at recombination junctions, in order to generate the repertoire of structurally diverse T cells necessary for antigen recognition. TCR beta subunits can be unambiguously identified by their hypervariable CDR3 (Complement Determining Region 3) sequence. This is the site of V(D)J recombination encoding the principal site of antigen contact. The complexity and dynamics of the T-cell repertoire remain unknown because the potential repertoire size has made conventional sequence analysis intractable. Here, we use 5'-RACE, Illumina sequencing, and a novel short read assembly strategy to sample CDR3(beta) diversity in human T lymphocytes from peripheral blood. Assembly of 40.5 million short reads identified 33,664 distinct TCR(beta) clonotypes and provides precise measurements of CDR3(beta) length diversity, usage of nontemplated bases, sequence convergence, and preferences for TRBV (T-cell receptor beta variable gene) and TRBJ (T-cell receptor beta joining gene) gene usage and pairing. CDR3 length between conserved residues of TRBV and TRBJ ranged from 21 to 81 nucleotides (nt). TRBV gene usage ranged from 0.01% for TRBV17 to 24.6% for TRBV20-1. TRBJ gene usage ranged from 1.6% for TRBJ2-6 to 17.2% for TRBJ2-1. We identified 1573 examples of convergence where the same amino acid translation was specified by distinct CDR3(beta) nucleotide sequences. Direct sequence-based immunoprofiling will likely prove to be a useful tool for understanding repertoire dynamics in response to immune challenge, without a priori knowledge of antigen.
Figures


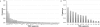
References
-
- Arstila TP, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286:958–961. - PubMed
-
- Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl.):S45–S55. - PubMed
-
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
