Antigen-specific proteolysis by hybrid antibodies containing promiscuous proteolytic light chains paired with an antigen-binding heavy chain
- PMID: 19542217
- PMCID: PMC2782051
- DOI: 10.1074/jbc.M109.011858
Antigen-specific proteolysis by hybrid antibodies containing promiscuous proteolytic light chains paired with an antigen-binding heavy chain
Abstract
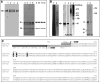
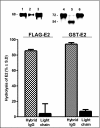
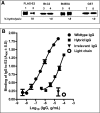
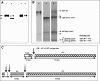
The antigen recognition site of antibodies consists of the heavy and light chain variable domains (V(L) and V(H) domains). V(L) domains catalyze peptide bond hydrolysis independent of V(H) domains (Mei, S., Mody, B., Eklund, S. H., and Paul, S. (1991) J. Biol. Chem. 266, 15571-15574). V(H) domains bind antigens noncovalently independent of V(L) domains (Ward, E. S., Güssow, D., Griffiths, A. D., Jones, P. T., and Winter, G. (1989) Nature 341, 544-546). We describe specific hydrolysis of fusion proteins of the hepatitis C virus E2 protein with glutathione S-transferase (GST-E2) or FLAG peptide (FLAG-E2) by antibodies containing the V(H) domain of an anti-E2 IgG paired with promiscuously catalytic V(L) domains. The hybrid IgG hydrolyzed the E2 fusion proteins more rapidly than the unpaired light chain. An active site-directed inhibitor of serine proteases inhibited the proteolytic activity of the hybrid IgG, indicating a serine protease mechanism. The hybrid IgG displayed noncovalent E2 binding in enzyme-linked immunosorbent assay tests. Immunoblotting studies suggested hydrolysis of FLAG-E2 at a bond within E2 located approximately 11 kDa from the N terminus. GST-E2 was hydrolyzed by the hybrid IgG at bonds in the GST tag. The differing cleavage pattern of FLAG-E2 and GST-E2 can be explained by the split-site model of catalysis, in which conformational differences in the E2 fusion protein substrates position alternate peptide bonds in register with the antibody catalytic subsite despite a common noncovalent binding mechanism. These studies provide proof-of-principle that the catalytic activity of a light chain can be rendered antigen-specific by pairing with a noncovalently binding heavy chain subunit.
Figures




Similar articles
-
Human recombinant antibodies specific for hepatitis C virus core and envelope E2 peptides from an immune phage display library.J Gen Virol. 1996 Oct;77 ( Pt 10):2531-9. doi: 10.1099/0022-1317-77-10-2531. J Gen Virol. 1996. PMID: 8887487
-
The antigen-binding domain of a human IgG-anti-F(ab')2 autoantibody.Proc Natl Acad Sci U S A. 1997 Mar 4;94(5):1902-7. doi: 10.1073/pnas.94.5.1902. Proc Natl Acad Sci U S A. 1997. PMID: 9050877 Free PMC article.
-
Expression and characterization of recombinant single-chain Fv and Fv fragments derived from a set of catalytic antibodies.Mol Immunol. 1997 Aug-Sep;34(12-13):891-906. doi: 10.1016/s0161-5890(97)00096-5. Mol Immunol. 1997. PMID: 9464525
-
Targeting random mutations to hotspots in antibody variable domains for affinity improvement.Methods Mol Biol. 2002;178:269-85. doi: 10.1385/1-59259-240-6:269. Methods Mol Biol. 2002. PMID: 11968497 Review. No abstract available.
-
IgG-like bispecific antibody platforms with built-in purification-facilitating elements.Protein Expr Purif. 2021 Dec;188:105955. doi: 10.1016/j.pep.2021.105955. Epub 2021 Aug 17. Protein Expr Purif. 2021. PMID: 34416361 Review.
Cited by
-
Catalytic Antibodies in Bipolar Disorder: Serum IgGs Hydrolyze Myelin Basic Protein.Int J Mol Sci. 2022 Jul 2;23(13):7397. doi: 10.3390/ijms23137397. Int J Mol Sci. 2022. PMID: 35806400 Free PMC article.
-
Specific amyloid β clearance by a catalytic antibody construct.J Biol Chem. 2015 Apr 17;290(16):10229-41. doi: 10.1074/jbc.M115.641738. Epub 2015 Feb 27. J Biol Chem. 2015. PMID: 25724648 Free PMC article.
-
Igg-Dependent Hydrolysis of Myelin Basic Protein of Patients with Different Courses of Schizophrenia.J Immunol Res. 2020 Aug 10;2020:8986521. doi: 10.1155/2020/8986521. eCollection 2020. J Immunol Res. 2020. PMID: 32851101 Free PMC article.
-
Biochemical features of a catalytic antibody light chain, 22F6, prepared from human lymphocytes.J Biol Chem. 2013 Jul 5;288(27):19558-68. doi: 10.1074/jbc.M113.454579. Epub 2013 May 15. J Biol Chem. 2013. PMID: 23677996 Free PMC article.
-
IgG Fab fragments forming bivalent complexes by a conformational mechanism that is reversible by osmolytes.J Biol Chem. 2012 Dec 14;287(51):42936-50. doi: 10.1074/jbc.M112.410217. Epub 2012 Oct 29. J Biol Chem. 2012. PMID: 23109335 Free PMC article.
References
-
- Sun M., Li L., Gao Q. S., Paul S. (1994) J. Biol. Chem. 269, 734–738 - PubMed
-
- Ward E. S., Güssow D., Griffiths A. D., Jones P. T., Winter G. (1989) Nature 341, 544–546 - PubMed
-
- Wilson I. A., Stanfield R. L., Rini J. M., Arevalo J. H., Schulze-Gahmen U., Fremont D. H., Stura E. A. (1991) CIBA Found. Symp. 159, 13–28; 28–39 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

