Structure-function relationships in miscoding by Sulfolobus solfataricus DNA polymerase Dpo4: guanine N2,N2-dimethyl substitution produces inactive and miscoding polymerase complexes
- PMID: 19542237
- PMCID: PMC2719408
- DOI: 10.1074/jbc.M109014274
Structure-function relationships in miscoding by Sulfolobus solfataricus DNA polymerase Dpo4: guanine N2,N2-dimethyl substitution produces inactive and miscoding polymerase complexes
Abstract
Previous work has shown that Y-family DNA polymerases tolerate large DNA adducts, but a substantial decrease in catalytic efficiency and fidelity occurs during bypass of N2,N2-dimethyl (Me2)-substituted guanine (N2,N2-Me2G), in contrast to a single methyl substitution. Therefore, it is unclear why the addition of two methyl groups is so disruptive. The presence of N2,N2-Me2G lowered the catalytic efficiency of the model enzyme Sulfolobus solfataricus Dpo4 16,000-fold. Dpo4 inserted dNTPs almost at random during bypass of N2,N2-Me2G, and much of the enzyme was kinetically trapped by an inactive ternary complex when N2,N2-Me2G was present, as judged by a reduced burst amplitude (5% of total enzyme) and kinetic modeling. One crystal structure of Dpo4 with a primer having a 3'-terminal dideoxycytosine (Cdd) opposite template N2,N2-Me2G in a post-insertion position showed Cdd folded back into the minor groove, as a catalytically incompetent complex. A second crystal had two unique orientations for the primer terminal Cdd as follows: (i) flipped into the minor groove and (ii) a long pairing with N2,N2-Me2G in which one hydrogen bond exists between the O-2 atom of Cdd and the N-1 atom of N2,N2-Me2G, with a second water-mediated hydrogen bond between the N-3 atom of Cdd and the O-6 atom of N2,N2-Me2G. A crystal structure of Dpo4 with dTTP opposite template N2,N2-Me2G revealed a wobble orientation. Collectively, these results explain, in a detailed manner, the basis for the reduced efficiency and fidelity of Dpo4-catalyzed bypass of N2,N2-Me2G compared with mono-substituted N2-alkyl G adducts.
Figures
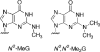




Similar articles
-
Recent insight into the kinetic mechanisms and conformational dynamics of Y-Family DNA polymerases.Biochemistry. 2014 May 6;53(17):2804-14. doi: 10.1021/bi5000405. Epub 2014 Apr 23. Biochemistry. 2014. PMID: 24716482 Free PMC article. Review.
-
Versatility of Y-family Sulfolobus solfataricus DNA polymerase Dpo4 in translesion synthesis past bulky N2-alkylguanine adducts.J Biol Chem. 2009 Feb 6;284(6):3563-76. doi: 10.1074/jbc.M807778200. Epub 2008 Dec 4. J Biol Chem. 2009. PMID: 19059910 Free PMC article.
-
Structural and functional analysis of Sulfolobus solfataricus Y-family DNA polymerase Dpo4-catalyzed bypass of the malondialdehyde-deoxyguanosine adduct.Biochemistry. 2009 Aug 4;48(30):7079-88. doi: 10.1021/bi9003588. Biochemistry. 2009. PMID: 19492857 Free PMC article.
-
Molecular basis of selectivity of nucleoside triphosphate incorporation opposite O6-benzylguanine by sulfolobus solfataricus DNA polymerase Dpo4: steady-state and pre-steady-state kinetics and x-ray crystallography of correct and incorrect pairing.J Biol Chem. 2007 May 4;282(18):13573-84. doi: 10.1074/jbc.M700656200. Epub 2007 Mar 3. J Biol Chem. 2007. PMID: 17337730
-
RB69 DNA polymerase structure, kinetics, and fidelity.Biochemistry. 2014 May 6;53(17):2752-67. doi: 10.1021/bi4014215. Epub 2014 Apr 23. Biochemistry. 2014. PMID: 24720884 Free PMC article. Review.
Cited by
-
Kinetic analysis of bypass of 7,8-dihydro-8-oxo-2'-deoxyguanosine by the catalytic core of yeast DNA polymerase η.Biochimie. 2016 Feb;121:161-9. doi: 10.1016/j.biochi.2015.12.009. Epub 2015 Dec 15. Biochimie. 2016. PMID: 26700143 Free PMC article.
-
Recent insight into the kinetic mechanisms and conformational dynamics of Y-Family DNA polymerases.Biochemistry. 2014 May 6;53(17):2804-14. doi: 10.1021/bi5000405. Epub 2014 Apr 23. Biochemistry. 2014. PMID: 24716482 Free PMC article. Review.
-
Noncognate DNA damage prevents the formation of the active conformation of the Y-family DNA polymerases DinB and DNA polymerase κ.FEBS J. 2015 Jul;282(14):2646-60. doi: 10.1111/febs.13304. Epub 2015 May 11. FEBS J. 2015. PMID: 25899385 Free PMC article.
-
Metal-ion dependence of the active-site conformation of the translesion DNA polymerase Dpo4 from Sulfolobus solfataricus.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Sep 1;66(Pt 9):1013-8. doi: 10.1107/S1744309110029374. Epub 2010 Aug 21. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010. PMID: 20823515 Free PMC article.
-
Mechanisms of mutagenesis: DNA replication in the presence of DNA damage.Mutat Res Rev Mutat Res. 2016 Apr-Jun;768:53-67. doi: 10.1016/j.mrrev.2016.03.006. Epub 2016 Apr 7. Mutat Res Rev Mutat Res. 2016. PMID: 27234563 Free PMC article. Review.
References
-
- Schaaper R. M., Glickman B. W., Loeb L. A. ( 1982) Cancer Res. 42, 3480– 3485 - PubMed
-
- Nakamura J., Walker V. E., Upton P. B., Chiang S. Y., Kow Y. W., Swenberg J. A. ( 1998) Cancer Res. 58, 222– 225 - PubMed
-
- Wink D. A., Kasprzak K. S., Maragos C. M., Elespuru R. K., Misra M., Dunams T. M., Cebula T. A., Koch W. H., Andrews A. W., Allen J. S., Keefer L. K. ( 1991) Science 254, 1001– 1003 - PubMed
-
- Kuchino Y., Mori F., Kasai H., Inoue H., Iwai S., Miura K., Ohtsuka E., Nishimura S. ( 1987) Nature 327, 77– 79 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

