Special Sm core complex functions in assembly of the U2 small nuclear ribonucleoprotein of Trypanosoma brucei
- PMID: 19542313
- PMCID: PMC2725569
- DOI: 10.1128/EC.00090-09
Special Sm core complex functions in assembly of the U2 small nuclear ribonucleoprotein of Trypanosoma brucei
Abstract
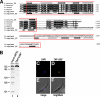
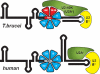
The processing of polycistronic pre-mRNAs in trypanosomes requires the spliceosomal small ribonucleoprotein complexes (snRNPs) U1, U2, U4/U6, U5, and SL, each of which contains a core of seven Sm proteins. Recently we reported the first evidence for a core variation in spliceosomal snRNPs; specifically, in the trypanosome U2 snRNP, two of the canonical Sm proteins, SmB and SmD3, are replaced by two U2-specific Sm proteins, Sm15K and Sm16.5K. Here we identify the U2-specific, nuclear-localized U2B'' protein from Trypanosoma brucei. U2B'' interacts with a second U2 snRNP protein, U2-40K (U2A'), which in turn contacts the U2-specific Sm16.5K/15K subcomplex. Together they form a high-affinity, U2-specific binding complex. This trypanosome-specific assembly differs from the mammalian system and provides a functional role for the Sm core variation found in the trypanosomal U2 snRNP.
Figures


Similar articles
-
Essential role of a trypanosome U4-specific Sm core protein in small nuclear ribonucleoprotein assembly and splicing.Eukaryot Cell. 2010 Mar;9(3):379-86. doi: 10.1128/EC.00353-09. Epub 2010 Jan 15. Eukaryot Cell. 2010. PMID: 20081062 Free PMC article.
-
Sm core variation in spliceosomal small nuclear ribonucleoproteins from Trypanosoma brucei.EMBO J. 2006 Oct 4;25(19):4513-23. doi: 10.1038/sj.emboj.7601328. Epub 2006 Sep 14. EMBO J. 2006. PMID: 16977313 Free PMC article.
-
Binding affinity and cooperativity control U2B″/snRNA/U2A' RNP formation.Biochemistry. 2014 Jun 17;53(23):3727-37. doi: 10.1021/bi500438e. Epub 2014 Jun 5. Biochemistry. 2014. PMID: 24866816 Free PMC article.
-
SMN-assisted assembly of snRNP-specific Sm cores in trypanosomes.Genes Dev. 2009 Jul 15;23(14):1650-64. doi: 10.1101/gad.526109. Genes Dev. 2009. PMID: 19605687 Free PMC article.
-
Assembly of the U2 small nuclear ribonucleoprotein from Trypanosoma brucei. A mutational analysis.J Biol Chem. 1993 Jun 25;268(18):13336-43. J Biol Chem. 1993. PMID: 8514772
Cited by
-
Genome-wide RNA-binding analysis of the trypanosome U1 snRNP proteins U1C and U1-70K reveals cis/trans-spliceosomal network.Nucleic Acids Res. 2014 Jun;42(10):6603-15. doi: 10.1093/nar/gku286. Epub 2014 Apr 19. Nucleic Acids Res. 2014. PMID: 24748659 Free PMC article.
-
Regulation of gene expression in trypanosomatids: living with polycistronic transcription.Open Biol. 2019 Jun 28;9(6):190072. doi: 10.1098/rsob.190072. Epub 2019 Jun 5. Open Biol. 2019. PMID: 31164043 Free PMC article.
-
Pseudouridines on Trypanosoma brucei spliceosomal small nuclear RNAs and their implication for RNA and protein interactions.Nucleic Acids Res. 2019 Aug 22;47(14):7633-7647. doi: 10.1093/nar/gkz477. Nucleic Acids Res. 2019. PMID: 31147702 Free PMC article.
-
The pre-mRNA splicing machinery of trypanosomes: complex or simplified?Eukaryot Cell. 2010 Aug;9(8):1159-70. doi: 10.1128/EC.00113-10. Epub 2010 Jun 25. Eukaryot Cell. 2010. PMID: 20581293 Free PMC article. Review.
-
Essential role of a trypanosome U4-specific Sm core protein in small nuclear ribonucleoprotein assembly and splicing.Eukaryot Cell. 2010 Mar;9(3):379-86. doi: 10.1128/EC.00353-09. Epub 2010 Jan 15. Eukaryot Cell. 2010. PMID: 20081062 Free PMC article.
References
-
- Berriman, M., et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309416-422. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

