Sensitive detection of chromosomal segments of distinct ancestry in admixed populations
- PMID: 19543370
- PMCID: PMC2689842
- DOI: 10.1371/journal.pgen.1000519
Sensitive detection of chromosomal segments of distinct ancestry in admixed populations
Abstract
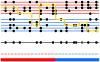
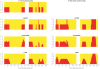
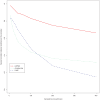
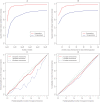
Identifying the ancestry of chromosomal segments of distinct ancestry has a wide range of applications from disease mapping to learning about history. Most methods require the use of unlinked markers; but, using all markers from genome-wide scanning arrays, it should in principle be possible to infer the ancestry of even very small segments with exquisite accuracy. We describe a method, HAPMIX, which employs an explicit population genetic model to perform such local ancestry inference based on fine-scale variation data. We show that HAPMIX outperforms other methods, and we explore its utility for inferring ancestry, learning about ancestral populations, and inferring dates of admixture. We validate the method empirically by applying it to populations that have experienced recent and ancient admixture: 935 African Americans from the United States and 29 Mozabites from North Africa. HAPMIX will be of particular utility for mapping disease genes in recently admixed populations, as its accurate estimates of local ancestry permit admixture and case-control association signals to be combined, enabling more powerful tests of association than with either signal alone.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
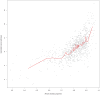
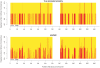
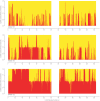
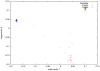
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

