Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus
- PMID: 19543527
- PMCID: PMC2694998
- DOI: 10.1371/journal.pone.0005660
Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus
Abstract
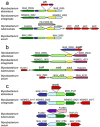
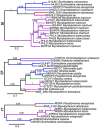
Mycobacterium abscessus is an emerging rapidly growing mycobacterium (RGM) causing a pseudotuberculous lung disease to which patients with cystic fibrosis (CF) are particularly susceptible. We report here its complete genome sequence. The genome of M. abscessus (CIP 104536T) consists of a 5,067,172-bp circular chromosome including 4920 predicted coding sequences (CDS), an 81-kb full-length prophage and 5 IS elements, and a 23-kb mercury resistance plasmid almost identical to pMM23 from Mycobacterium marinum. The chromosome encodes many virulence proteins and virulence protein families absent or present in only small numbers in the model RGM species Mycobacterium smegmatis. Many of these proteins are encoded by genes belonging to a "mycobacterial" gene pool (e.g. PE and PPE proteins, MCE and YrbE proteins, lipoprotein LpqH precursors). However, many others (e.g. phospholipase C, MgtC, MsrA, ABC Fe(3+) transporter) appear to have been horizontally acquired from distantly related environmental bacteria with a high G+C content, mostly actinobacteria (e.g. Rhodococcus sp., Streptomyces sp.) and pseudomonads. We also identified several metabolic regions acquired from actinobacteria and pseudomonads (relating to phenazine biosynthesis, homogentisate catabolism, phenylacetic acid degradation, DNA degradation) not present in the M. smegmatis genome. Many of the "non mycobacterial" factors detected in M. abscessus are also present in two of the pathogens most frequently isolated from CF patients, Pseudomonas aeruginosa and Burkholderia cepacia. This study elucidates the genetic basis of the unique pathogenicity of M. abscessus among RGM, and raises the question of similar mechanisms of pathogenicity shared by unrelated organisms in CF patients.
Conflict of interest statement
Figures



Similar articles
-
Genome analysis reveals three genomospecies in Mycobacterium abscessus.BMC Genomics. 2014 May 12;15(1):359. doi: 10.1186/1471-2164-15-359. BMC Genomics. 2014. PMID: 24886480 Free PMC article.
-
Functional characterization of the Mycobacterium abscessus genome coupled with condition specific transcriptomics reveals conserved molecular strategies for host adaptation and persistence.BMC Genomics. 2016 Aug 5;17:553. doi: 10.1186/s12864-016-2868-y. BMC Genomics. 2016. PMID: 27495169 Free PMC article.
-
Mycobacterium abscessus phospholipase C expression is induced during coculture within amoebae and enhances M. abscessus virulence in mice.Infect Immun. 2015 Feb;83(2):780-91. doi: 10.1128/IAI.02032-14. Epub 2014 Dec 8. Infect Immun. 2015. PMID: 25486995 Free PMC article.
-
Mycobacterium abscessus, a complex of three fast-growing subspecies sharing virulence traits with slow-growing mycobacteria.Clin Microbiol Infect. 2024 Jun;30(6):726-731. doi: 10.1016/j.cmi.2023.08.036. Epub 2023 Oct 4. Clin Microbiol Infect. 2024. PMID: 37797823 Review.
-
Mycobacterial Pathogenomics and Evolution.Microbiol Spectr. 2014 Feb;2(1):MGM2-0025-2013. doi: 10.1128/microbiolspec.MGM2-0025-2013. Microbiol Spectr. 2014. PMID: 26082120 Review.
Cited by
-
Pipeline of anti-Mycobacterium abscessus small molecules: Repurposable drugs and promising novel chemical entities.Med Res Rev. 2021 Jul;41(4):2350-2387. doi: 10.1002/med.21798. Epub 2021 Mar 1. Med Res Rev. 2021. PMID: 33645845 Free PMC article. Review.
-
Evaluation of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Identification of Mycobacterium abscessus Subspecies According to Whole-Genome Sequencing.J Clin Microbiol. 2016 Dec;54(12):2982-2989. doi: 10.1128/JCM.01151-16. Epub 2016 Sep 28. J Clin Microbiol. 2016. PMID: 27682129 Free PMC article.
-
Comparative functional pan-genome analyses to build connections between genomic dynamics and phenotypic evolution in polycyclic aromatic hydrocarbon metabolism in the genus Mycobacterium.BMC Evol Biol. 2015 Feb 14;15:21. doi: 10.1186/s12862-015-0302-8. BMC Evol Biol. 2015. PMID: 25880171 Free PMC article.
-
Omadacycline for management of Mycobacterium abscessus infections: a review of its effectiveness, place in therapy, and considerations for use.BMC Infect Dis. 2022 Nov 22;22(1):874. doi: 10.1186/s12879-022-07857-7. BMC Infect Dis. 2022. PMID: 36419143 Free PMC article. Review.
-
Distribution of horizontally transferred heavy metal resistance operons in recent outbreak bacteria.Mob Genet Elements. 2012 Mar 1;2(2):96-100. doi: 10.4161/mge.19963. Mob Genet Elements. 2012. PMID: 22934243 Free PMC article.
References
-
- Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175:367–416. - PubMed
-
- Moore M, Frerichs JB. An unusual acid-fast infection of the knee with subcutaneous, abscess-like lesions of the gluteal region; report of a case with a study of the organism, Mycobacterium abscessus, n. sp. J Invest Dermatol. 1953;20:133–169. - PubMed
-
- Kubica GP, Baess I, Gordon RE, Jenkins PA, Kwapinski JB, et al. A co-operative numerical analysis of rapidly growing mycobacteria. J Gen Microbiol. 1972;73:55–70. - PubMed
-
- Kusunoki S, Ezaki T. Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status: Mycobacterium abscessus comb. nov. Int J Syst Bacteriol. 1992;42:240–245. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

