ES cell cycle progression and differentiation require the action of the histone methyltransferase Dot1L
- PMID: 19544450
- PMCID: PMC6338332
- DOI: 10.1002/stem.86
ES cell cycle progression and differentiation require the action of the histone methyltransferase Dot1L
Abstract
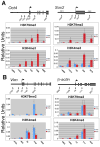
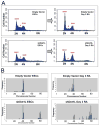
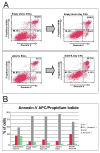
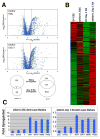
Mouse embryonic stem cells (ESCs) proliferate with rapid cell cycle kinetics but without loss of pluripotency. The histone methyltransferase Dot1L is responsible for methylation of histone H3 at lysine 79 (H3K79me). We investigated whether ESCs require Dot1L for proper stem cell behavior. ESCs deficient in Dot1L tolerate a nearly complete loss of H3K79 methylation without a substantial impact on proliferation or morphology. However, shortly after differentiation is induced, Dot1L-deficient cells cease proliferating and arrest in G2/M-phase of the cell cycle, with increased levels of aneuploidy. In addition, many aberrant mitotic spindles occur in Dot1L-deficient cells. Surprisingly, these mitotic and cell cycle defects fail to trigger apoptosis, indicating that mouse ESCs lack stringent cell cycle checkpoint control during initial stages of differentiation. Transcriptome analysis indicates that Dot1L deficiency causes the misregulation of a select set of genes, including many with known roles in cell cycle control and cellular proliferation as well as markers of endoderm differentiation. The data indicate a requirement for Dot1L function for early stages of ESC differentiation where Dot1L is necessary for faithful execution of mitosis and proper transcription of many genes throughout the genome.
Figures


References
-
- Rasmussen T. Developmentally-poised chromatin of embryonic stem cells. Front Biosci. 2008;13:1568–1577. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. - PubMed
-
- Feldman N, Gerson A, Fang J, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8(2):188–194. - PubMed
-
- Feng Q, Wang H, Ng HH, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12(12):1052–1058. - PubMed
-
- Krogan NJ, Dover J, Wood A, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11(3):721–729. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

