Effect of protein structure on deamidation rate in the Fc fragment of an IgG1 monoclonal antibody
- PMID: 19544580
- PMCID: PMC2776945
- DOI: 10.1002/pro.173
Effect of protein structure on deamidation rate in the Fc fragment of an IgG1 monoclonal antibody
Abstract
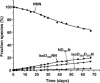
The effects of secondary structure on asparagine (N) deamidation in a 22 amino acid sequence (369-GFYPSDIAVEWESNGQPENNYK-390) of the crystallizable (Fc) fragment of a human monoclonal antibody (Fc IgG1) were investigated using high-resolution ultra performance liquid chromatography with tandem mass spectrometry (UPLC/MS). Samples containing either the intact Fc IgG (approximately 50 kD) ("intact protein"), or corresponding synthetic peptides ("peptide") were stored in Tris buffer at 37 degrees C and pH 7.5 for up to forty days, then subjected to UPLC/MS analysis with high energy MS1 fragmentation. The peptide deamidated only at N(382) to form the isoaspartate (isoD(382)) and aspartate (D(382)) products in the ratio of approximately 4:1, with a half-life of approximately 3.4 days. The succinimide intermediate (Su(382)) was also detected; deamidation was not observed for the other two sites (N(387) and N(388)) in peptide samples. The intact protein showed a 30-fold slower overall deamidation half-life of approximately 108 days to produce the isoD(382) and D(387) products, together with minor amounts of D(382). Surprisingly, the D(382) and isoD(387) products were not detected in intact protein samples and, as in the peptide samples, deamidation was not detected at N(388). The results indicate that higher order structure influences both the rate of N-deamidation and the product distribution.
Figures


 ,
,  , and
, and  ions, together with the elution and fragmentation patterns of synthetic peptide standards (see text), identifies the peaks as the IsoD382NN, NNN (parent) and D382NN forms, respectively. See the electronic version of this article for enlarged figures.
ions, together with the elution and fragmentation patterns of synthetic peptide standards (see text), identifies the peaks as the IsoD382NN, NNN (parent) and D382NN forms, respectively. See the electronic version of this article for enlarged figures.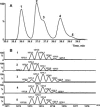


Similar articles
-
Characterization of asparagine 330 deamidation in an Fc-fragment of IgG1 using cation exchange chromatography and peptide mapping.J Chromatogr B Analyt Technol Biomed Life Sci. 2014 Aug 15;965:65-71. doi: 10.1016/j.jchromb.2014.06.018. Epub 2014 Jun 21. J Chromatogr B Analyt Technol Biomed Life Sci. 2014. PMID: 24999246
-
Characterization of IgG1 Fc Deamidation at Asparagine 325 and Its Impact on Antibody-dependent Cell-mediated Cytotoxicity and FcγRIIIa Binding.Sci Rep. 2020 Jan 15;10(1):383. doi: 10.1038/s41598-019-57184-2. Sci Rep. 2020. PMID: 31941950 Free PMC article.
-
Asparagine deamidation dependence on buffer type, pH, and temperature.J Pharm Sci. 2013 Jun;102(6):1712-1723. doi: 10.1002/jps.23529. Epub 2013 Apr 9. J Pharm Sci. 2013. PMID: 23568760
-
Profiling the 'deamidome' of complex biosamples using mixed-mode chromatography-coupled tandem mass spectrometry.Methods. 2022 Apr;200:31-41. doi: 10.1016/j.ymeth.2020.05.005. Epub 2020 May 8. Methods. 2022. PMID: 32418626 Review.
-
Functional Role of Carbohydrate Residues in Human Immunoglobulin G and Therapeutic Monoclonal Antibodies.Biochemistry (Mosc). 2016 Aug;81(8):835-57. doi: 10.1134/S0006297916080058. Biochemistry (Mosc). 2016. PMID: 27677552 Review.
Cited by
-
Risk-Based Control Strategies of Recombinant Monoclonal Antibody Charge Variants.Antibodies (Basel). 2022 Nov 20;11(4):73. doi: 10.3390/antib11040073. Antibodies (Basel). 2022. PMID: 36412839 Free PMC article. Review.
-
Analytical comparability study of recombinant monoclonal antibody therapeutics.MAbs. 2018 May/Jun;10(4):513-538. doi: 10.1080/19420862.2018.1438797. Epub 2018 Mar 20. MAbs. 2018. PMID: 29513619 Free PMC article. Review.
-
Kinetic, thermodynamic, and ab initio insights of AsnGly isomerisation as a ticking time bomb for protein integrity.Commun Chem. 2024 Dec 19;7(1):303. doi: 10.1038/s42004-024-01374-1. Commun Chem. 2024. PMID: 39702829 Free PMC article.
-
Characterization of therapeutic monoclonal antibodies reveals differences between in vitro and in vivo time-course studies.Pharm Res. 2013 Jan;30(1):167-78. doi: 10.1007/s11095-012-0860-z. Epub 2012 Sep 6. Pharm Res. 2013. PMID: 22956170
-
Enzymatic attributes of an l-isoaspartyl methyltransferase from Candida utilis and its role in cell survival.Biochem Biophys Rep. 2015 Aug 28;4:59-75. doi: 10.1016/j.bbrep.2015.08.015. eCollection 2015 Dec. Biochem Biophys Rep. 2015. PMID: 29124188 Free PMC article.
References
-
- Chowdhury PS, Wu H. Tailor-made antibody therapeutics. Methods. 2005;36:11–24. - PubMed
-
- Gottschalk U. Downstream processing of monoclonal antibodies: from high dilution to high purity. Bio Pharm Int. 2005;18:42–44. 46, 48, 50, 52, 54, 56, 58.
-
- Aswad DW. Deamidation and isoaspartate formation in peptides and proteins. Ann Arbor: CRC Press; 1995. p. 259.
-
- Jenkins N, Murphy L, Tyther R. Post-translational modifications of recombinant proteins: significance for biopharmaceuticals. Mol Biotechnol. 2008;39:113–118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

