Hypoxia-inducible factor prolyl-4-hydroxylase PHD2 protein abundance depends on integral membrane anchoring of FKBP38
- PMID: 19546213
- PMCID: PMC2755711
- DOI: 10.1074/jbc.M109.032631
Hypoxia-inducible factor prolyl-4-hydroxylase PHD2 protein abundance depends on integral membrane anchoring of FKBP38
Abstract
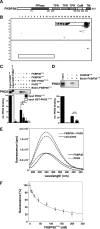
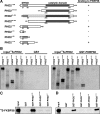
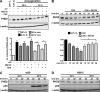
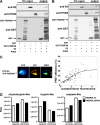
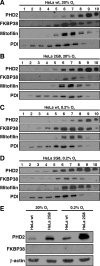
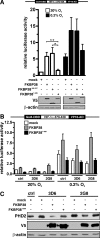
Prolyl-4-hydroxylase domain (PHD) proteins are 2-oxoglutarate and dioxygen-dependent enzymes that mediate the rapid destruction of hypoxia-inducible factor alpha subunits. Whereas PHD1 and PHD3 proteolysis has been shown to be regulated by Siah2 ubiquitin E3 ligase-mediated polyubiquitylation and proteasomal destruction, protein regulation of the main oxygen sensor responsible for hypoxia-inducible factor alpha regulation, PHD2, remained unknown. We recently reported that the FK506-binding protein (FKBP) 38 specifically interacts with PHD2 and determines PHD2 protein stability in a peptidyl-prolyl cis-trans isomerase-independent manner. Using peptide array binding assays, fluorescence spectroscopy, and fluorescence resonance energy transfer analysis, we defined a minimal linear glutamate-rich PHD2 binding domain in the N-terminal part of FKBP38 and showed that this domain forms a high affinity complex with PHD2. Vice versa, PHD2 interacted with a non-linear N-terminal motif containing the MYND (myeloid, Nervy, and DEAF-1)-type Zn(2+) finger domain with FKBP38. Biochemical fractionation and immunofluorescence analysis demonstrated that PHD2 subcellular localization overlapped with FKBP38 in the endoplasmic reticulum and mitochondria. An additional fraction of PHD2 was found in the cytoplasm. In cellulo PHD2/FKBP38 association, as well as regulation of PHD2 protein abundance by FKBP38, is dependent on membrane- anchored FKBP38 localization mediated by the C-terminal transmembrane domain. Mechanistically our data indicate that PHD2 protein stability is regulated by a ubiquitin-independent proteasomal pathway involving FKBP38 as adaptor protein that mediates proteasomal interaction. We hypothesize that FKBP38-bound PHD2 is constantly degraded whereas cytosolic PHD2 is stable and able to function as an active prolyl-4-hydroxylase.
Figures

References
-
- Semenza G. L. (2007) Sci. STKE 2007,cm8. - PubMed
-
- Wenger R. H., Stiehl D. P., Camenisch G. (2005) Sci. STKE 2005,re12. - PubMed
-
- Bruick R. K., McKnight S. L. (2001) Science 294,1337–1340 - PubMed
-
- Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) Cell 107,43–54 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

