Prion protein-detergent micelle interactions studied by NMR in solution
- PMID: 19546219
- PMCID: PMC2755680
- DOI: 10.1074/jbc.M109.000430
Prion protein-detergent micelle interactions studied by NMR in solution
Abstract
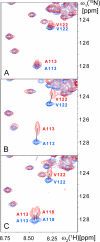
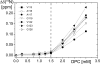
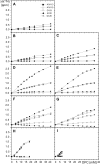
Cellular prion proteins, PrP(C), carrying the amino acid substitutions P102L, P105L, or A117V, which confer increased susceptibility to human transmissible spongiform encephalopathies, are known to form structures that include transmembrane polypeptide segments. Herein, we investigated the interactions between dodecylphosphocholine micelles and the polypeptide fragments 90-231 of the recombinant mouse PrP variants carrying the amino acid replacements P102L, P105L, A117V, A113V/A115V/A118V, K110I/H111I, M129V, P105L/M129V, and A117V/M129V. Wild-type mPrP-(90-231) and mPrP[M129V]-(91-231) showed only weak interactions with dodecylphosphocholine micelles in aqueous solution at pH 7.0, whereas discrete interaction sites within the polypeptide segment 102-127 were identified for all other aforementioned mPrP variants by NMR chemical shift mapping. These model studies thus provide evidence that amino acid substitutions within the polypeptide segment 102-127 affect the interactions of PrP(C) with membranous structures, which might in turn modulate the physiological function of the protein in health and disease.
Figures



Similar articles
-
Prion protein NMR structure from tammar wallaby (Macropus eugenii) shows that the beta2-alpha2 loop is modulated by long-range sequence effects.J Mol Biol. 2009 Jun 26;389(5):833-45. doi: 10.1016/j.jmb.2009.04.040. Epub 2009 Apr 23. J Mol Biol. 2009. PMID: 19393664
-
Interactions between the conserved hydrophobic region of the prion protein and dodecylphosphocholine micelles.J Biol Chem. 2012 Jan 13;287(3):1915-22. doi: 10.1074/jbc.M111.279364. Epub 2011 Nov 29. J Biol Chem. 2012. PMID: 22128151 Free PMC article.
-
Prion protein NMR structure and familial human spongiform encephalopathies.Proc Natl Acad Sci U S A. 1998 Sep 29;95(20):11667-72. doi: 10.1073/pnas.95.20.11667. Proc Natl Acad Sci U S A. 1998. PMID: 9751723 Free PMC article.
-
Molecular dynamics studies on 3D structures of the hydrophobic region PrP(109-136).Acta Biochim Biophys Sin (Shanghai). 2013 Jun;45(6):509-19. doi: 10.1093/abbs/gmt031. Epub 2013 Apr 5. Acta Biochim Biophys Sin (Shanghai). 2013. PMID: 23563221 Review.
-
Perturbations of Native Membrane Protein Structure in Alkyl Phosphocholine Detergents: A Critical Assessment of NMR and Biophysical Studies.Chem Rev. 2018 Apr 11;118(7):3559-3607. doi: 10.1021/acs.chemrev.7b00570. Epub 2018 Feb 28. Chem Rev. 2018. PMID: 29488756 Free PMC article. Review.
Cited by
-
Structural Determinants of the Prion Protein N-Terminus and Its Adducts with Copper Ions.Int J Mol Sci. 2018 Dec 20;20(1):18. doi: 10.3390/ijms20010018. Int J Mol Sci. 2018. PMID: 30577569 Free PMC article. Review.
-
Residues within a lipid-associated segment of the PECAM-1 cytoplasmic domain are susceptible to inducible, sequential phosphorylation.Blood. 2011 Jun 2;117(22):6012-23. doi: 10.1182/blood-2010-11-317867. Epub 2011 Apr 4. Blood. 2011. PMID: 21464369 Free PMC article.
-
Copper alters aggregation behavior of prion protein and induces novel interactions between its N- and C-terminal regions.J Biol Chem. 2011 Nov 4;286(44):38533-38545. doi: 10.1074/jbc.M111.265645. Epub 2011 Sep 7. J Biol Chem. 2011. PMID: 21900252 Free PMC article.
-
Structural Basis for Ca2+-mediated Interaction of the Perforin C2 Domain with Lipid Membranes.J Biol Chem. 2015 Oct 16;290(42):25213-26. doi: 10.1074/jbc.M115.668384. Epub 2015 Aug 25. J Biol Chem. 2015. PMID: 26306037 Free PMC article.
-
Rapid ex vivo reverse genetics identifies the essential determinants of prion protein toxicity.Brain Pathol. 2023 Mar;33(2):e13130. doi: 10.1111/bpa.13130. Epub 2022 Nov 3. Brain Pathol. 2023. PMID: 36329611 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

