A randomly integrated transgenic H19 imprinting control region acquires methylation imprinting independently of its establishment in germ cells
- PMID: 19546235
- PMCID: PMC2725707
- DOI: 10.1128/MCB.00275-09
A randomly integrated transgenic H19 imprinting control region acquires methylation imprinting independently of its establishment in germ cells
Abstract
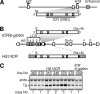
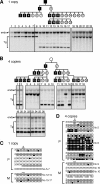
The imprinted expression of the mouse Igf2/H19 locus is governed by the differential methylation of the imprinting control region (ICR), which is established initially in germ cells and subsequently maintained in somatic cells, depending on its parental origin. By grafting a 2.9-kbp H19 ICR fragment into a human beta-globin yeast artificial chromosome in transgenic mice, we previously showed that the ICR could recapitulate imprinted methylation and expression at a heterologous locus, suggesting that the H19 ICR in the beta-globin locus contained sufficient information to maintain the methylation mark (K. Tanimoto, M. Shimotsuma, H. Matsuzaki, A. Omori, J. Bungert, J. D. Engel, and A. Fukamizu, Proc. Natl. Acad. Sci. USA 102:10250-10255, 2005). Curiously, however, the transgenic H19 ICR was not methylated in sperm, which was distinct from that seen in the endogenous locus. Here, we reevaluated the ability of the H19 ICR to mark the parental origin using more rigid criteria. In the testis, the methylation levels of the solitary 2.9-kbp transgenic ICR fragment varied significantly between six transgenic mouse lines. However, in somatic cells, the paternally inherited ICR fragment exhibited consistently higher methylation levels at five out of six randomly integrated sites in the mouse genome. These results clearly demonstrated that the H19 ICR could acquire parent-of-origin-dependent methylation after fertilization independently of the chromosomal integration site or the prerequisite methylation acquisition in male germ cells.
Figures



References
-
- Ainscough, J. F., T. Koide, M. Tada, S. Barton, and M. A. Surani. 1997. Imprinting of Igf2 and H19 from a 130 kb YAC transgene. Development 1243621-3632. - PubMed
-
- Bartolomei, M. S., S. Zemel, and S. M. Tilghman. 1991. Parental imprinting of the mouse H19 gene. Nature 351153-155. - PubMed
-
- Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405482-485. - PubMed
-
- Birger, Y., R. Shemer, J. Perk, and A. Razin. 1999. The imprinting box of the mouse Igf2r gene. Nature 39784-88. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
