MURC/Cavin-4 and cavin family members form tissue-specific caveolar complexes
- PMID: 19546242
- PMCID: PMC2712963
- DOI: 10.1083/jcb.200903053
MURC/Cavin-4 and cavin family members form tissue-specific caveolar complexes
Abstract
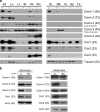
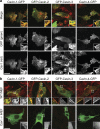
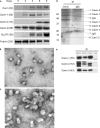
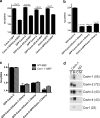
Polymerase I and transcript release factor (PTRF)/Cavin is a cytoplasmic protein whose expression is obligatory for caveola formation. Using biochemistry and fluorescence resonance energy transfer-based approaches, we now show that a family of related proteins, PTRF/Cavin-1, serum deprivation response (SDR)/Cavin-2, SDR-related gene product that binds to C kinase (SRBC)/Cavin-3, and muscle-restricted coiled-coil protein (MURC)/Cavin-4, forms a multiprotein complex that associates with caveolae. This complex can constitutively assemble in the cytosol and associate with caveolin at plasma membrane caveolae. Cavin-1, but not other cavins, can induce caveola formation in a heterologous system and is required for the recruitment of the cavin complex to caveolae. The tissue-restricted expression of cavins suggests that caveolae may perform tissue-specific functions regulated by the composition of the cavin complex. Cavin-4 is expressed predominantly in muscle, and its distribution is perturbed in human muscle disease associated with Caveolin-3 dysfunction, identifying Cavin-4 as a novel muscle disease candidate caveolar protein.
Figures



References
-
- Birnbaum M.J. 1989. Identification of a novel gene encoding an insulin-responsive glucose transporter protein.Cell. 57:305–315 - PubMed
-
- Capozza F., Combs T.P., Cohen A.W., Cho Y.R., Park S.Y., Schubert W., Williams T.M., Brasaemle D.L., Jelicks L.A., Scherer P.E., et al. 2005. Caveolin-3 knockout mice show increased adiposity and whole body insulin resistance, with ligand-induced insulin receptor instability in skeletal muscle.Am. J. Physiol. Cell Physiol. 288:C1317–C1331 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

