Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy
- PMID: 19547746
- PMCID: PMC2691949
- DOI: 10.1371/journal.pbio.1000137
Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy
Abstract
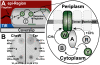
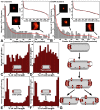
The Escherichia coli chemotaxis network is a model system for biological signal processing. In E. coli, transmembrane receptors responsible for signal transduction assemble into large clusters containing several thousand proteins. These sensory clusters have been observed at cell poles and future division sites. Despite extensive study, it remains unclear how chemotaxis clusters form, what controls cluster size and density, and how the cellular location of clusters is robustly maintained in growing and dividing cells. Here, we use photoactivated localization microscopy (PALM) to map the cellular locations of three proteins central to bacterial chemotaxis (the Tar receptor, CheY, and CheW) with a precision of 15 nm. We find that cluster sizes are approximately exponentially distributed, with no characteristic cluster size. One-third of Tar receptors are part of smaller lateral clusters and not of the large polar clusters. Analysis of the relative cellular locations of 1.1 million individual proteins (from 326 cells) suggests that clusters form via stochastic self-assembly. The super-resolution PALM maps of E. coli receptors support the notion that stochastic self-assembly can create and maintain approximately periodic structures in biological membranes, without direct cytoskeletal involvement or active transport.
Conflict of interest statement
The authors declare competing financial interests. EB and Harald Hess (Janelia Farm) have licensed the PALM technology to Carl Zeiss Microimaging, GmbH.
Figures


References
-
- Adler J. Chemotaxis in bacteria. Annu Rev Biochem. 1975;44:341–356. - PubMed
-
- Kim KK, Yokota H, Kim S-H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. - PubMed
-
- Maddock J, Shapiro L. Polar localization of the chemoreceptor complex in the E. coli cell. Science. 1993;259:1717–1723. - PubMed
-
- Baker M, Wolanin P, Stock J. Signal transduction in bacterial chemotaxis. BioEssays. 2006;28:9–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

