Hepatitis C virus core, NS3, NS4B and NS5A are the major immunogenic proteins in humoral immunity in chronic HCV infection
- PMID: 19549310
- PMCID: PMC2709157
- DOI: 10.1186/1743-422X-6-84
Hepatitis C virus core, NS3, NS4B and NS5A are the major immunogenic proteins in humoral immunity in chronic HCV infection
Abstract
Background: The viral genome of hepatitis C virus constitutes a 9.6-kb single-stranded positive-sense RNA which encodes altogether 11 viral proteins. In order to study the humoral immune responses against different HCV proteins in patients suffering from chronic HCV infection, we produced three structural (core, E1 and E2) and six nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A and NS5B) in Sf9 insect cells by using the baculovirus expression system.
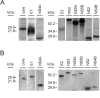
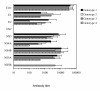
Results: The recombinant HCV core, E1, E2, NS2, NS3, NS4A, NS4B, NS5A and NS5B proteins were purified and used in Western blot analysis to determine antibody responses against individual HCV protein in 68 HCV RNA and antibody positive human sera that were obtained from patients suffering from genotype 1, 2, 3 or 4 infection. These sera were also analysed with INNO-LIA Score test for HCV antibodies against core, NS3, NS4AB and NS5A, and the results were similar to the ones obtained by Western blot method. Based on our Western blot analyses we found that the major immunogenic HCV antigens were the core, NS4B, NS3 and NS5A proteins which were recognized in 97%, 86%, 68% and 53% of patient sera, respectively. There were no major genotype specific differences in antibody responses to individual HCV proteins. A common feature within the studied sera was that all except two sera recognized the core protein in high titers, whereas none of the sera recognized NS2 protein and only three sera (from genotype 3) recognised NS5B.
Conclusion: The data shows significant variation in the specificity in humoral immunity in chronic HCV patients.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

