Polypeptide translocation by the AAA+ ClpXP protease machine
- PMID: 19549599
- PMCID: PMC2718738
- DOI: 10.1016/j.chembiol.2009.05.007
Polypeptide translocation by the AAA+ ClpXP protease machine
Abstract
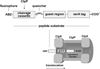
In the AAA+ ClpXP protease, ClpX uses repeated cycles of ATP hydrolysis to pull native proteins apart and to translocate the denatured polypeptide into ClpP for degradation. Here, we probe polypeptide features important for translocation. ClpXP degrades diverse synthetic peptide substrates despite major differences in side-chain chirality, size, and polarity. Moreover, translocation occurs without a peptide -NH and with 10 methylenes between successive peptide bonds. Pulling on homopolymeric tracts of glycine, proline, and lysine also allows efficient ClpXP degradation of a stably folded protein. Thus, minimal chemical features of a polypeptide chain are sufficient for translocation and protein unfolding by the ClpX machine. These results suggest that the translocation pore of ClpX is highly elastic, allowing interactions with a wide range of chemical groups, a feature likely to be shared by many AAA+ unfoldases.
Figures
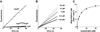


References
-
- Adzhubei AA, Sternberg MJE. Left-handed polyproline II helices commonly occur in globular proteins. J. Mol. Biol. 1993;229:472–493. - PubMed
-
- Bolon DN, Grant RA, Baker TA, Sauer RT. Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol. Cell. 2004;16:343–350. - PubMed
-
- Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

