Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis
- PMID: 19549685
- PMCID: PMC2704886
- DOI: 10.1242/jcs.046250
Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis
Abstract
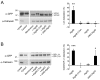
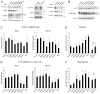
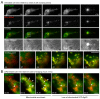
Autophagy is an important catabolic process with roles in cell survival and cell death. It sequesters cytosol and organelles within double-membrane autophagosomes that deliver their contents to lysosomes for degradation. Autophagosome biogenesis is coordinated by the autophagy-related protein 4 (Atg4) family of C54 endopeptidases (Atg4A-Atg4D). These enzymes prime and then later delipidate the autophagosome marker, Atg8. Here, we show that one family member, Atg4D, is cleaved by caspase-3 in vitro and in apoptotic cells. Atg4D is a poor priming and delipidation enzyme in vitro, but truncated DeltaN63 Atg4D displays increased activity against the Atg8 paralogue, gamma-aminobutyric acid receptor-associated protein-like 1 (GABARAP-L1). In living cells, DeltaN63 Atg4D stimulates the delipidation of GABARAP-L1, whereas siRNA silencing of the gene expressing Atg4D abrogates GABARAP-L1 autophagosome formation and sensitises cells to starvation and staurosporine-induced cell death. Interestingly, Atg4D overexpression induces apoptosis, which is preceded by the caspase-independent recruitment of Atg4D to mitochondria and is facilitated by a putative C-terminal Bcl-2 homology 3 (BH3) domain. Atg4D also acquires affinity for damaged mitochondria in cells treated with hydrogen peroxide. These data suggest that Atg4D is an autophagy regulator that links mitochondrial dysfunction with apoptosis.
Figures







References
-
- Axe, E. L., Walker, S. A., Manifava, M., Chandra, P., Roderick, H. L., Habermann, A., Griffiths, G. and Ktistakis, N. T. (2008). Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685-701. - PMC - PubMed
-
- Baehrecke, E. H. (2005). Autophagy: dual roles in life and death? Nat. Rev. Mol. Cell Biol. 6, 505-510. - PubMed
-
- Botti, J., Djavaheri-Mergny, M., Pilatte, Y. and Codogno, P. (2006). Autophagy signaling and the cogwheels of cancer. Autophagy 2, 67-73. - PubMed
-
- Broustas, C. G., Gokhale, P. C., Rahman, A., Dritschilo, A., Ahmad, I. and Kasid, U. (2004). BRCC2, a novel BH3-like domain-containing protein, induces apoptosis in a caspase-dependent manner. J. Biol. Chem. 279, 26780-26788. - PubMed
-
- Chen, G., Cizeau, J., Vande Velde, C., Park, J. H., Bozek, G., Bolton, J., Shi, L., Dubik, D. and Greenberg, A. (1999). Nix and Nip3 form a subfamily of pro-apoptotic mitochondrial proteins. J. Biol. Chem. 274, 7-10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

