Suppression of nonhomologous end joining repair by overexpression of HMGA2
- PMID: 19549901
- PMCID: PMC2737594
- DOI: 10.1158/0008-5472.CAN-08-4833
Suppression of nonhomologous end joining repair by overexpression of HMGA2
Abstract
Understanding the molecular details associated with aberrant high mobility group A2 (HMGA2) gene expression is key to establishing the mechanism(s) underlying its oncogenic potential and effect on the development of therapeutic strategies. Here, we report the involvement of HMGA2 in impairing DNA-dependent protein kinase (DNA-PK) during the nonhomologous end joining (NHEJ) process. We showed that HMGA2-expressing cells displayed deficiency in overall and precise DNA end-joining repair and accumulated more endogenous DNA damage. Proper and timely activation of DNA-PK, consisting of Ku70, Ku80, and DNA-PKcs subunits, is essential for the repair of DNA double strand breaks (DSB) generated endogenously or by exposure to genotoxins. In cells overexpressing HMGA2, accumulation of histone 2A variant X phosphorylation at Ser-139 (gamma-H2AX) was associated with hyperphosphorylation of DNA-PKcs at Thr-2609 and Ser-2056 before and after the induction of DSBs. Also, the steady-state complex of Ku and DNA ends was altered by HMGA2. Microirradiation and real-time imaging in living cells revealed that HMGA2 delayed the release of DNA-PKcs from DSB sites, similar to observations found in DNA-PKcs mutants. Moreover, HMGA2 alone was sufficient to induce chromosomal aberrations, a hallmark of deficiency in NHEJ-mediated DNA repair. In summary, a novel role for HMGA2 to interfere with NHEJ processes was uncovered, implicating HMGA2 in the promotion of genome instability and tumorigenesis.
Figures
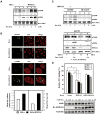

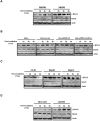
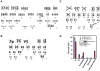
Comment in
-
Comment re: HMGA2 is a negative regulator of DNA-PK pathway.Cancer Res. 2010 Feb 15;70(4):1742; author reply 1742. doi: 10.1158/0008-5472.CAN-09-3081. Epub 2010 Feb 9. Cancer Res. 2010. PMID: 20145117 No abstract available.
References
-
- Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions (Review) Int J Oncol. 2008;32:289–305. - PubMed
-
- Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–4. - PubMed
-
- Monzen K, Ito Y, Naito AT, et al. A crucial role of a high mobility group protein HMGA2 in cardiogenesis. Nat Cell Biol. 2008;10:567–74. - PubMed
-
- Fedele M, Visone R, De Martino I, et al. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

