DNA methylation regulates constitutive expression of Stat6 regulatory genes SOCS-1 and SHP-1 in colon cancer cells
- PMID: 19551406
- PMCID: PMC11844784
- DOI: 10.1007/s00432-009-0627-z
DNA methylation regulates constitutive expression of Stat6 regulatory genes SOCS-1 and SHP-1 in colon cancer cells
Abstract
Purpose: Stat6 signaling is active in cancer cells and IL-4-induced Stat6 activities or Stat6 activational phenotypes vary among cancer cells. This study aimed at investigating possible mechanism(s) involved in the formation of varying Stat6 activities/phenotypes.
Methods: Stat6 regulatory genes, SOCS-1 and SHP-1, were examined for mRNA expression using RT-PCR, and their promoter DNA methylation was assayed by methylation-specific PCR in Stat6-phenotyped colon cancer cell lines. DNA methylation was then verified by sequencing. RT-PCR assay and Western blotting were used to detect the expression of SOCS-1 and SHP-1 after demethylation using 5-aza-2'-deoxycytidine.
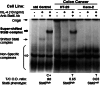
Results: Compared with Stat6(null) Caco-2 cells, Stat6(high) HT-29 cells showed decreased constitutive expression of SOCS-1 and SHP-1, which correlated with DNA hypermethylation in these genes' promoters. Interestingly, demethylation in HT-29 cells recovered the constitutive expression of SOCS-1 and SHP-1.
Conclusions: These findings suggest that DNA methylation controls the constitutive expression of negative Stat6 regulatory genes, which may affect Stat6 activities.
Figures




Similar articles
-
Differential IL-4/Stat6 activities correlate with differential expression of regulatory genes SOCS-1, SHP-1, and PP2A in colon cancer cells.J Cancer Res Clin Oncol. 2009 Jan;135(1):131-40. doi: 10.1007/s00432-008-0429-8. Epub 2008 Jun 7. J Cancer Res Clin Oncol. 2009. PMID: 18536936 Free PMC article.
-
Defective interleukin-4/Stat6 activity correlates with increased constitutive expression of negative regulators SOCS-3, SOCS-7, and CISH in colon cancer cells.J Interferon Cytokine Res. 2009 Dec;29(12):809-16. doi: 10.1089/jir.2009.0004. J Interferon Cytokine Res. 2009. PMID: 19929568
-
Frequent methylation of RASSF1A in synovial sarcoma and the anti-tumor effects of 5-aza-2'-deoxycytidine against synovial sarcoma cell lines.J Cancer Res Clin Oncol. 2010 Jan;136(1):17-25. doi: 10.1007/s00432-009-0632-2. J Cancer Res Clin Oncol. 2010. PMID: 19578875 Free PMC article.
-
Clinical and biological effects of demethylating agents on solid tumours - A systematic review.Cancer Treat Rev. 2017 Mar;54:10-23. doi: 10.1016/j.ctrv.2017.01.004. Epub 2017 Jan 18. Cancer Treat Rev. 2017. PMID: 28189913
-
Epigenetic silencing of the tumor suppressor TIMP-3 gene in upstream CpG islands in thyroid neoplasms: a cross-sectional study with systematic review.Mol Biol Rep. 2025 Jul 1;52(1):665. doi: 10.1007/s11033-025-10721-x. Mol Biol Rep. 2025. PMID: 40593331
Cited by
-
Immune markers and differential signaling networks in ulcerative colitis and Crohn's disease.Inflamm Bowel Dis. 2012 Dec;18(12):2342-56. doi: 10.1002/ibd.22957. Epub 2012 Mar 29. Inflamm Bowel Dis. 2012. PMID: 22467146 Free PMC article.
-
The role of suppressors of cytokine signalling in human neoplasms.Mol Biol Int. 2014;2014:630797. doi: 10.1155/2014/630797. Epub 2014 Mar 16. Mol Biol Int. 2014. PMID: 24757565 Free PMC article. Review.
-
Role and mechanism of decitabine combined with tyrosine kinase inhibitors in advanced chronic myeloid leukemia cells.Oncol Lett. 2017 Aug;14(2):1295-1302. doi: 10.3892/ol.2017.6318. Epub 2017 Jun 6. Oncol Lett. 2017. PMID: 28789344 Free PMC article.
-
Effects of Herba epimedii and Fructus ligustri lucidi on the transcription factors in hypothalamus of aged rats.Chin J Integr Med. 2011 Oct;17(10):758-63. doi: 10.1007/s11655-011-0636-z. Epub 2011 Mar 30. Chin J Integr Med. 2011. PMID: 21465296
-
Constitutive Activation of Interleukin-13/STAT6 Contributes to Kaposi's Sarcoma-Associated Herpesvirus-Related Primary Effusion Lymphoma Cell Proliferation and Survival.J Virol. 2015 Oct;89(20):10416-26. doi: 10.1128/JVI.01525-15. Epub 2015 Aug 5. J Virol. 2015. PMID: 26246572 Free PMC article.
References
-
- Ansel KM, Djuretic I, Tanasa B, Rao A (2006) Regulation of Th2 differentiation and IL-4 locus accessibility. Annu Rev Immunol 24:607–656. doi:10.1146/annurev.immunol.23.021704.115821 - PubMed
-
- Baylin S (2001) DNA methylation and epigenetic mechanisms of carcinogenesis. Dev Biol (Basel) 106:85–87 - PubMed
-
- Bruns HA, Kaplan MH (2006) The role of constitutively active Stat6 in leukemia and lymphoma. Crit Rev Oncol Hematol 57(3):245–253. doi:10.1016/j.critrevonc.2005.08.005 - PubMed
-
- Dickensheets H, Vazquez N, Sheikh F et al (2007) Suppressor of cytokine signaling-1 is an IL-4-inducible gene in macrophages and feedback inhibits IL-4 signaling. Genes Immun 8:21–27. doi:10.1038/sj.gene.6364352 - PubMed
-
- Esteller M, Corn PG, Baylin SB, Herman JG (2001) A gene hypermethylation profile of human cancer. Cancer Res 61:3225–3229 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

