Mixed infection and the genesis of influenza virus diversity
- PMID: 19553313
- PMCID: PMC2738154
- DOI: 10.1128/JVI.00773-09
Mixed infection and the genesis of influenza virus diversity
Abstract
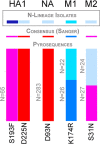
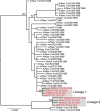
The emergence of viral infections with potentially devastating consequences for human health is highly dependent on their underlying evolutionary dynamics. One likely scenario for an avian influenza virus, such as A/H5N1, to evolve to one capable of human-to-human transmission is through the acquisition of genetic material from the A/H1N1 or A/H3N2 subtypes already circulating in human populations. This would require that viruses of both subtypes coinfect the same cells, generating a mixed infection, and then reassort. Determining the nature and frequency of mixed infection with influenza virus is therefore central to understanding the emergence of pandemic, antigenic, and drug-resistant strains. To better understand the potential for such events, we explored patterns of intrahost genetic diversity in recently circulating strains of human influenza virus. By analyzing multiple viral genome sequences sampled from individual influenza patients we reveal a high level of mixed infection, including diverse lineages of the same influenza virus subtype, drug-resistant and -sensitive strains, those that are likely to differ in antigenicity, and even viruses of different influenza virus types (A and B). These results reveal that individuals can harbor influenza viruses that differ in major phenotypic properties, including those that are antigenically distinct and those that differ in their sensitivity to antiviral agents.
Figures



References
-
- Centers for Disease Control and Prevention. 1998. Update: influenza activity—United States and worldwide, 1997-98 season, and composition of the 1998-99 influenza vaccine. MMWR Morb. Mortal. Wkly. Rep. 47280-284. - PubMed
-
- Delcher, A. L., S. L. Salzberg, and A. M. Phillippy. 2003. Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinformatics 1010.3. - PubMed
-
- Dugan, V. G., R. Chen, D. J. Spiro, N. Sengamalay, J. Zaborsky, E. Ghedin, J. Nolting, D. E. Swayne, J. A. Runstadler, G. M. Happ, D. A. Senne, R. Wang, R. D. Slemons, E. C. Holmes, and J. K. Taubenberger. 2008. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 4e1000076. - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

