Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells
- PMID: 19553314
- PMCID: PMC2738192
- DOI: 10.1128/JVI.00613-09
Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells
Abstract
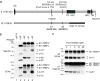
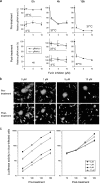
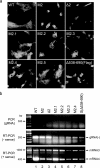
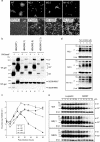
The spike (S) protein of the coronavirus (CoV) infectious bronchitis virus (IBV) is cleaved into S1 and S2 subunits at the furin consensus motif RRFRR(537)/S in virus-infected cells. In this study, we observe that the S2 subunit of the IBV Beaudette strain is additionally cleaved at the second furin site (RRRR(690)/S) in cells expressing S constructs and in virus-infected cells. Detailed time course experiments showed that a peptide furin inhibitor, decanoyl-Arg-Val-Lys-Arg-chloromethylketone, blocked both viral entry and syncytium formation. Site-directed mutagenesis studies revealed that the S1/S2 cleavage by furin was not necessary for, but could promote, syncytium formation by and infectivity of IBV in Vero cells. In contrast, the second site is involved in the furin dependence of viral entry and syncytium formation. Mutations of the second site from furin-cleavable RRRR/S to non-furin-cleavable PRRRS and AAARS, respectively, abrogated the furin dependence of IBV entry. Instead, a yet-to-be-identified serine protease(s) was involved, as revealed by protease inhibitor studies. Furthermore, sequence analysis of CoV S proteins by multiple alignments showed conservation of an XXXR/S motif, cleavable by either furin or other trypsin-like proteases, at a position equivalent to the second IBV furin site. Taken together, these results suggest that proteolysis at a novel XXXR/S motif in the S2 subunit might be a common mechanism for the entry of CoV into cells.
Figures



Similar articles
-
D614G Substitution of SARS-CoV-2 Spike Protein Increases Syncytium Formation and Virus Titer via Enhanced Furin-Mediated Spike Cleavage.mBio. 2021 Aug 31;12(4):e0058721. doi: 10.1128/mBio.00587-21. Epub 2021 Jul 27. mBio. 2021. PMID: 34311586 Free PMC article.
-
Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites.Proc Natl Acad Sci U S A. 2009 Apr 7;106(14):5871-6. doi: 10.1073/pnas.0809524106. Epub 2009 Mar 24. Proc Natl Acad Sci U S A. 2009. PMID: 19321428 Free PMC article.
-
The S2 Subunit of Infectious Bronchitis Virus Beaudette Is a Determinant of Cellular Tropism.J Virol. 2018 Sep 12;92(19):e01044-18. doi: 10.1128/JVI.01044-18. Print 2018 Oct 1. J Virol. 2018. PMID: 30021894 Free PMC article.
-
[Protease-dependent cell entry mechanism of coronaviruses].Uirusu. 2011 Jun;61(1):109-16. doi: 10.2222/jsv.61.109. Uirusu. 2011. PMID: 21972562 Review. Japanese.
-
Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence.Viruses. 2012 Apr;4(4):557-80. doi: 10.3390/v4040557. Epub 2012 Apr 12. Viruses. 2012. PMID: 22590686 Free PMC article. Review.
Cited by
-
Overcoming Culture Restriction for SARS-CoV-2 in Human Cells Facilitates the Screening of Compounds Inhibiting Viral Replication.Antimicrob Agents Chemother. 2021 Jun 17;65(7):e0009721. doi: 10.1128/AAC.00097-21. Epub 2021 Jun 17. Antimicrob Agents Chemother. 2021. PMID: 33903110 Free PMC article.
-
Alternative Antigen Processing for MHC Class I: Multiple Roads Lead to Rome.Front Immunol. 2015 Jun 5;6:298. doi: 10.3389/fimmu.2015.00298. eCollection 2015. Front Immunol. 2015. PMID: 26097483 Free PMC article. Review.
-
Human coronavirus HKU1 infection of primary human type II alveolar epithelial cells: cytopathic effects and innate immune response.PLoS One. 2013 Jul 24;8(7):e70129. doi: 10.1371/journal.pone.0070129. Print 2013. PLoS One. 2013. PMID: 23894604 Free PMC article.
-
Kidney Injury in COVID-19: Epidemiology, Molecular Mechanisms and Potential Therapeutic Targets.Int J Mol Sci. 2022 Feb 17;23(4):2242. doi: 10.3390/ijms23042242. Int J Mol Sci. 2022. PMID: 35216358 Free PMC article. Review.
-
Proteases facilitate the endosomal escape of porcine epidemic diarrhea virus during entry into host cells.Virus Res. 2019 Oct 15;272:197730. doi: 10.1016/j.virusres.2019.197730. Epub 2019 Aug 21. Virus Res. 2019. PMID: 31445102 Free PMC article.
References
-
- Basak, A., M. Zhong, J. S. Munzer, M. Chretien, and N. G. Seidah. 2001. Implication of the proprotein convertases furin, PC5 and PC7 in the cleavage of surface glycoproteins of Hong Kong, Ebola and respiratory syncytial viruses: a comparative analysis with fluorogenic peptides. Biochem. J. 353537-545. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

