Interaction of CDK5RAP2 with EB1 to track growing microtubule tips and to regulate microtubule dynamics
- PMID: 19553473
- PMCID: PMC2777926
- DOI: 10.1091/mbc.e09-01-0009
Interaction of CDK5RAP2 with EB1 to track growing microtubule tips and to regulate microtubule dynamics
Abstract
Mutations in cdk5rap2 are linked to autosomal recessive primary microcephaly, and attention has been paid to its function at centrosomes. In this report, we demonstrate that CDK5RAP2 localizes to microtubules and concentrates at the distal tips in addition to centrosomal localization. CDK5RAP2 interacts directly with EB1, a prototypic member of microtubule plus-end tracking proteins, and contains the basic and Ser-rich motif responsible for EB1 binding. The EB1-binding motif is conserved in the CDK5RAP2 sequences of chimpanzee, bovine, and dog but not in those of rat and mouse, suggesting a function gained during the evolution of mammals. The mutation of the Ile/Leu-Pro dipeptide within the motif abolishes EB1 interaction and plus-end attachment. In agreement with the mutational analysis, suppression of EB1 expression inhibits microtubule tip-tracking of CDK5RAP2. We have also found that the CDK5RAP2-EB1 complex regulates microtubule dynamics and stability. CDK5RAP2 depletion by RNA interference impacts the dynamic behaviors of microtubules. The CDK5RAP2-EB1 complex induces microtubule bundling and acetylation when expressed in cell cultures and stimulates microtubule assembly and bundle formation in vitro. Collectively, these results show that CDK5RAP2 targets growing microtubule tips in association with EB1 to regulate microtubule dynamics.
Figures

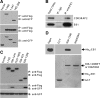

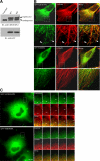
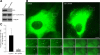
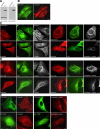
References
-
- Akhmanova A., Steinmetz M. O. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 2008;9:309–322. - PubMed
-
- Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. - PubMed
-
- Askham J. M., Moncur P., Markham A. F., Morrison E. E. Regulation and function of the interaction between the APC tumour suppressor protein and EB1. Oncogene. 2000;19:1950–1958. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

