A progesterone receptor co-activator (JDP2) mediates activity through interaction with residues in the carboxyl-terminal extension of the DNA binding domain
- PMID: 19553667
- PMCID: PMC2782034
- DOI: 10.1074/jbc.M109.003244
A progesterone receptor co-activator (JDP2) mediates activity through interaction with residues in the carboxyl-terminal extension of the DNA binding domain
Abstract
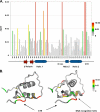
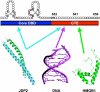
Progesterone receptor (PR) belongs to the nuclear receptor family of ligand-dependent transcription factors and mediates the major biological effects of progesterone. Transcriptional co-activators that are recruited by PR through the carboxyl-terminal ligand binding domain have been studied extensively. Much less is known about co-activators that interact with other regions of receptors. Jun dimerization protein 2 (JDP2) is a PR co-activator that enhances the transcriptional activity of the amino-terminal domain by increasing the alpha-helical content and stability of the intrinsically disordered amino-terminal domain. To gain insights into the mechanism of JDP2 co-activation of PR, the structural basis of JDP2-PR interaction was analyzed using NMR. The smallest regions of each protein needed for efficient protein interaction were used for NMR and included the basic region plus leucine zipper (bZIP) domain of JDP2 and the core zinc modules of the PR DNA binding domain plus the intrinsically disordered carboxyl-terminal extension (CTE) of the DNA binding domain. Chemical shift changes in PR upon titration with JDP2 revealed that most of the residues involved in binding of JDP2 reside within the CTE. The importance of the CTE for binding JDP2 was confirmed by peptide competition and mutational analyses. Point mutations within CTE sites identified by NMR and a CTE domain swapping experiment also confirmed the functional importance of JDP2 interaction with the CTE for enhancement of PR transcriptional activity. These studies provide insights into the role and functional importance of the CTE for co-activator interactions.
Figures



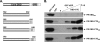

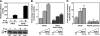
Similar articles
-
Regulation of the amino-terminal transcription activation domain of progesterone receptor by a cofactor-induced protein folding mechanism.Mol Cell Biol. 2005 Oct;25(20):8792-808. doi: 10.1128/MCB.25.20.8792-8808.2005. Mol Cell Biol. 2005. PMID: 16199860 Free PMC article.
-
Jun dimerization protein 2 functions as a progesterone receptor N-terminal domain coactivator.Mol Cell Biol. 2002 Aug;22(15):5451-66. doi: 10.1128/MCB.22.15.5451-5466.2002. Mol Cell Biol. 2002. PMID: 12101239 Free PMC article.
-
Mechanism of high-mobility group protein B enhancement of progesterone receptor sequence-specific DNA binding.Nucleic Acids Res. 2008 Jun;36(11):3655-66. doi: 10.1093/nar/gkn249. Epub 2008 May 12. Nucleic Acids Res. 2008. PMID: 18474528 Free PMC article.
-
Structural and functional analysis of domains of the progesterone receptor.Mol Cell Endocrinol. 2012 Jan 30;348(2):418-29. doi: 10.1016/j.mce.2011.07.017. Epub 2011 Jul 22. Mol Cell Endocrinol. 2012. PMID: 21803119 Free PMC article. Review.
-
Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action.Recent Prog Horm Res. 1997;52:141-64; discussion 164-5. Recent Prog Horm Res. 1997. PMID: 9238851 Review.
Cited by
-
Multiple functions of the histone chaperone Jun dimerization protein 2.Gene. 2016 Sep 30;590(2):193-200. doi: 10.1016/j.gene.2016.03.048. Epub 2016 Apr 8. Gene. 2016. PMID: 27041241 Free PMC article. Review.
-
Binding of the N-terminal region of coactivator TIF2 to the intrinsically disordered AF1 domain of the glucocorticoid receptor is accompanied by conformational reorganizations.J Biol Chem. 2012 Dec 28;287(53):44546-60. doi: 10.1074/jbc.M112.411330. Epub 2012 Nov 6. J Biol Chem. 2012. PMID: 23132854 Free PMC article.
-
PRMT1 Is Critical for the Transcriptional Activity and the Stability of the Progesterone Receptor.iScience. 2020 Jun 26;23(6):101236. doi: 10.1016/j.isci.2020.101236. Epub 2020 Jun 4. iScience. 2020. PMID: 32563156 Free PMC article.
-
Ligand binding domain mutations of the glucocorticoid receptor selectively modify the effects with, but not binding of, cofactors.Biochemistry. 2011 Jan 25;50(3):356-66. doi: 10.1021/bi101792d. Epub 2010 Dec 30. Biochemistry. 2011. PMID: 21142156 Free PMC article.
-
Dynamic transcriptome, accessible genome, and PGR cistrome profiles in the human myometrium.FASEB J. 2020 Feb;34(2):2252-2268. doi: 10.1096/fj.201902654R. Epub 2019 Dec 12. FASEB J. 2020. PMID: 31908010 Free PMC article.
References
-
- Chawla A., Repa J. J., Evans R. M., Mangelsdorf D. J. (2001) Science 294, 1866–1870 - PubMed
-
- Li X., O'Malley B. W. (2003) J. Biol. Chem. 278, 39261–39264 - PubMed
-
- Meyer M. E., Quirin-Stricker C., Lerouge T., Bocquel M. T., Gronemeyer H. (1992) J. Biol. Chem. 267, 10882–10887 - PubMed
-
- Bourguet W., Germain P., Gronemeyer H. (2000) Trends Pharmacol. Sci. 21, 381–388 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

