GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer
- PMID: 19553991
- PMCID: PMC2753613
- DOI: 10.1038/nature08109
GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer
Abstract
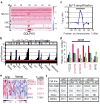
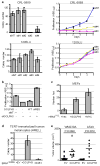
Genome-wide copy number analyses of human cancers identified a frequent 5p13 amplification in several solid tumour types, including lung (56%), ovarian (38%), breast (32%), prostate (37%) and melanoma (32%). Here, using integrative analysis of a genomic profile of the region, we identify a Golgi protein, GOLPH3, as a candidate targeted for amplification. Gain- and loss-of-function studies in vitro and in vivo validated GOLPH3 as a potent oncogene. Physically, GOLPH3 localizes to the trans-Golgi network and interacts with components of the retromer complex, which in yeast has been linked to target of rapamycin (TOR) signalling. Mechanistically, GOLPH3 regulates cell size, enhances growth-factor-induced mTOR (also known as FRAP1) signalling in human cancer cells, and alters the response to an mTOR inhibitor in vivo. Thus, genomic and genetic, biological, functional and biochemical data in yeast and humans establishes GOLPH3 as a new oncogene that is commonly targeted for amplification in human cancer, and is capable of modulating the response to rapamycin, a cancer drug in clinical use.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

