Fine-scale dissection of functional protein network organization by statistical network analysis
- PMID: 19554104
- PMCID: PMC2699632
- DOI: 10.1371/journal.pone.0006017
Fine-scale dissection of functional protein network organization by statistical network analysis
Abstract
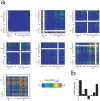
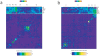
Revealing organizational principles of biological networks is an important goal of systems biology. In this study, we sought to analyze the dynamic organizational principles within the protein interaction network by studying the characteristics of individual neighborhoods of proteins within the network based on their gene expression as well as protein-protein interaction patterns. By clustering proteins into distinct groups based on their neighborhood gene expression characteristics, we identify several significant trends in the dynamic organization of the protein interaction network. We show that proteins with distinct neighborhood gene expression characteristics are positioned in specific localities in the protein interaction network thereby playing specific roles in the dynamic network connectivity. Remarkably, our analysis reveals a neighborhood characteristic that corresponds to the most centrally located group of proteins within the network. Further, we show that the connectivity pattern displayed by this group is consistent with the notion of "rich club connectivity" in complex networks. Importantly, our findings are largely reproducible in networks constructed using independent and different datasets.
Conflict of interest statement
Figures

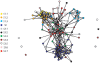
Similar articles
-
Network legos: building blocks of cellular wiring diagrams.J Comput Biol. 2008 Sep;15(7):829-44. doi: 10.1089/cmb.2007.0139. J Comput Biol. 2008. PMID: 18707557
-
Comparative network analysis via differential graphlet communities.Proteomics. 2015 Jan;15(2-3):608-17. doi: 10.1002/pmic.201400233. Epub 2014 Dec 15. Proteomics. 2015. PMID: 25283527 Free PMC article.
-
Biological impacts and context of network theory.J Exp Biol. 2007 May;210(Pt 9):1548-58. doi: 10.1242/jeb.003731. J Exp Biol. 2007. PMID: 17449819 Review.
-
Revealing static and dynamic modular architecture of the eukaryotic protein interaction network.Mol Syst Biol. 2007;3:110. doi: 10.1038/msb4100149. Epub 2007 Apr 24. Mol Syst Biol. 2007. PMID: 17453049 Free PMC article.
-
Dynamic proteomics in modeling of the living cell. Protein-protein interactions.Biochemistry (Mosc). 2009 Dec;74(13):1586-607. doi: 10.1134/s0006297909130112. Biochemistry (Mosc). 2009. PMID: 20210711 Review.
Cited by
-
Patterns of human gene expression variance show strong associations with signaling network hierarchy.BMC Syst Biol. 2010 Nov 12;4:154. doi: 10.1186/1752-0509-4-154. BMC Syst Biol. 2010. PMID: 21073694 Free PMC article.
-
Bioinformatics and systems biology.Mol Oncol. 2012 Apr;6(2):147-54. doi: 10.1016/j.molonc.2012.01.008. Epub 2012 Feb 17. Mol Oncol. 2012. PMID: 22377422 Free PMC article. Review.
-
Generation of 2-mode scale-free graphs for link-level internet topology modeling.PLoS One. 2020 Nov 9;15(11):e0240100. doi: 10.1371/journal.pone.0240100. eCollection 2020. PLoS One. 2020. PMID: 33166286 Free PMC article.
References
-
- Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. - PubMed
-
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. - PubMed
-
- Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. - PubMed
-
- Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. - PubMed
-
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, et al. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

