Sp4-dependent repression of neurotrophin-3 limits dendritic branching
- PMID: 19555762
- PMCID: PMC2727929
- DOI: 10.1016/j.mcn.2009.06.008
Sp4-dependent repression of neurotrophin-3 limits dendritic branching
Abstract
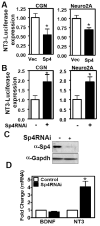
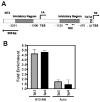
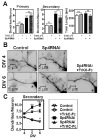
Regulation of neuronal gene expression is critical to establish functional connections in the mammalian nervous system. The transcription factor Sp4 regulates dendritic patterning during cerebellar granule neuron development by limiting branching and promoting activity-dependent pruning. Here, we investigate neurotrophin-3 (NT3) as a target gene important for Sp4-dependent dendritic morphogenesis. We found that Sp4 overexpression reduced NT3 promoter activity whereas knockdown of Sp4 increased NT3 promoter activity and mRNA. Moreover, Sp4 bound to the NT3 promoter in vivo, supporting a direct role for Sp4 as a repressor of NT3 expression. Addition of exogenous NT3 promoted dendritic branching in cerebellar granule neurons. Furthermore, sequestering NT3 blocked the continued addition of dendritic branches observed upon Sp4 knockdown, but had no effect on dendrite pruning. These findings demonstrate that, during cerebellar granule neuron development, Sp4-dependent repression of neurotrophin-3 is required to limit dendritic branching and thereby promote acquisition of the mature dendritic pattern.
Figures


Similar articles
-
Transcription factor Sp4 regulates dendritic patterning during cerebellar maturation.Proc Natl Acad Sci U S A. 2007 Jun 5;104(23):9882-7. doi: 10.1073/pnas.0701946104. Epub 2007 May 29. Proc Natl Acad Sci U S A. 2007. PMID: 17535924 Free PMC article.
-
Transcription factor Sp4 regulates expression of nervous wreck 2 to control NMDAR1 levels and dendrite patterning.Dev Neurobiol. 2015 Jan;75(1):93-108. doi: 10.1002/dneu.22212. Epub 2014 Jul 22. Dev Neurobiol. 2015. PMID: 25045015 Free PMC article.
-
Phosphorylation of the transcription factor Sp4 is reduced by NMDA receptor signaling.J Neurochem. 2014 May;129(4):743-52. doi: 10.1111/jnc.12657. Epub 2014 Feb 12. J Neurochem. 2014. PMID: 24475768 Free PMC article.
-
Functional roles of neurotrophin 3 in the developing and mature sympathetic nervous system.Mol Neurobiol. 1996 Dec;13(3):185-97. doi: 10.1007/BF02740622. Mol Neurobiol. 1996. PMID: 8989769 Review.
-
Neurotrophin-3 in the development of the enteric nervous system.Prog Brain Res. 2004;146:243-63. doi: 10.1016/S0079-6123(03)46016-0. Prog Brain Res. 2004. PMID: 14699968 Review.
Cited by
-
Store-operated calcium entry promotes the degradation of the transcription factor Sp4 in resting neurons.Sci Signal. 2014 Jun 3;7(328):ra51. doi: 10.1126/scisignal.2005242. Sci Signal. 2014. PMID: 24894994 Free PMC article.
-
Transcriptional Regulators in the Cerebellum in Chronic Schizophrenia: Novel Possible Targets for Pharmacological Interventions.Int J Mol Sci. 2025 Apr 12;26(8):3653. doi: 10.3390/ijms26083653. Int J Mol Sci. 2025. PMID: 40332239 Free PMC article.
-
Environmental enrichment reverses cerebellar impairments caused by prenatal exposure to a synthetic glucocorticoid.AIMS Neurosci. 2022 Jul 14;9(3):320-344. doi: 10.3934/Neuroscience.2022018. eCollection 2022. AIMS Neurosci. 2022. PMID: 36329900 Free PMC article.
-
Transcription factor SP4 phosphorylation is altered in the postmortem cerebellum of bipolar disorder and schizophrenia subjects.Eur Neuropsychopharmacol. 2015 Oct;25(10):1650-1660. doi: 10.1016/j.euroneuro.2015.05.006. Epub 2015 May 21. Eur Neuropsychopharmacol. 2015. PMID: 26049820 Free PMC article.
-
Transcriptional regulation of neuronal polarity and morphogenesis in the mammalian brain.Neuron. 2011 Oct 6;72(1):22-40. doi: 10.1016/j.neuron.2011.09.018. Neuron. 2011. PMID: 21982366 Free PMC article. Review.
References
-
- Altman J. Postnatal development of the cerebellar cortex in the rat. 3. Maturation of the components of the granular layer. J Comp Neurol. 1972;145:465–513. - PubMed
-
- Baker RE, Dijkhuizen PA, Van Pelt J, Verhaagen J. Growth of pyramidal, but not non-pyramidal, dendrites in long-term organotypic explants of neonatal rat neocortex chronically exposed to neurotrophin-3. Eur J Neurosci. 1998;10:1037–1044. - PubMed
-
- Bates B, Rios M, Trumpp A, Chen C, Fan G, Bishop JM, Jaenisch R. Neurotrophin-3 is required for proper cerebellar development. Nat Neurosci. 1999;2:115–117. - PubMed
-
- Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. - PubMed
-
- Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

