Identification of a maize neocentromere in an oat-maize addition line
- PMID: 19556776
- PMCID: PMC2813801
- DOI: 10.1159/000218128
Identification of a maize neocentromere in an oat-maize addition line
Abstract
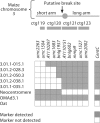
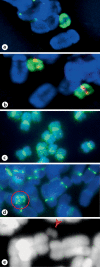
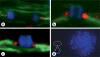
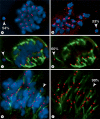
We report a neocentromere event on maize chromosome 3 that occurred due to chromosome breakage. The neocentromere lies on a fragment of the short arm that lacks the primary centromere DNA elements, CentC and CRM. It is transmitted in the genomic background of oat via a new centromere (and kinetochore), as shown by immunolocalization of the oat CENH3 protein. Despite normal transmission of the maize fragment in most progeny, neocentromeres appear to vary in size within the same tissue, as shown by fluorescent measurements. A secondary truncation in one line lowered mitotic transmission to 3% and precipitously reduced the size of the chromosome. The results support the view that neocentromere formation is generally associated with major genomic disturbances such as wide species crosses or deletion of an existing centromere. The data further suggest that new centromeres may undergo a period of instability that is corrected over a period of several generations.
(c) 2009 S. Karger AG, Basel.
Figures



References
-
- Alonso A, Mahmood R, Li S, Cheung F, Yoda K, Warburton PE. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32. Hum Mol Genet. 2003;12:2711–2721. - PubMed
-
- Bhattacharyya GK, Johnson RA. Statistical Concepts and Methods. New York: John Wiley & Sons; 1977. p. 604.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

