Cell-autonomous requirement for DNaseII in nonapoptotic cell death
- PMID: 19557011
- PMCID: PMC2770252
- DOI: 10.1038/cdd.2009.79
Cell-autonomous requirement for DNaseII in nonapoptotic cell death
Abstract
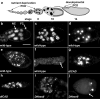
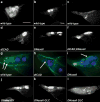
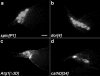
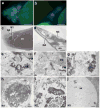
DNA fragmentation is a critical component of apoptosis but it has not been characterized in nonapoptotic forms of cell death, such as necrosis and autophagic cell death. In mammalian apoptosis, caspase-activated DNase cleaves DNA into nucleosomal fragments in dying cells, and subsequently DNase II, an acid nuclease, completes the DNA degradation but acts non-cell autonomously within lysosomes of engulfing cells. Here we examine the requirement for DNases during two examples of programmed cell death (PCD) that occurs in the Drosophila melanogaster ovary, starvation-induced death of mid-stage egg chambers and developmental nurse cell death in late oogenesis. Surprisingly, we found that DNaseII was required cell autonomously in nurse cells during developmental PCD, indicating that it acts within dying cells. Dying nurse cells contain autophagosomes, indicating that autophagy may contribute to these forms of PCD. Furthermore, we provide evidence that developmental nurse cell PCD in late oogenesis shows hallmarks of necrosis. These findings indicate that DNaseII can act cell autonomously to degrade DNA during nonapoptotic cell death.
Figures

References
-
- Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–254. - PubMed
-
- Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. - PubMed
-
- Nagata S. DNA degradation in development and programmed cell death. Annu Rev Immunol. 2005;23:853–875. - PubMed
-
- Parrish JZ, Xue D. Cuts can kill: the roles of apoptotic nucleases in cell death and animal development. Chromosoma. 2006;115:89–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

